-
PDF
- Split View
-
Views
-
Cite
Cite
Susan E. Taymans, Svetlana Pack, Evgenia Pak, Zsolt Orban, Julia Barsony, Zhengping Zhuang, Constantine A. Stratakis, The Human Vitamin D Receptor Gene (VDR) Is Localized to Region 12cen‐q12 by Fluorescent In Situ Hybridization and Radiation Hybrid Mapping: Genetic and Physical VDR Map, Journal of Bone and Mineral Research, Volume 14, Issue 7, 1 July 1999, Pages 1163–1166, https://doi.org/10.1359/jbmr.1999.14.7.1163
Close - Share Icon Share
Abstract
The vitamin D receptor (VDR) is a member of the steroid hormone receptor superfamily of ligand‐activated transcription factors. The VDR gene was previously mapped to human chromosome 12q13–12q14, but its precise physical and genetic localization are unknown. The present study reports the mapping of the human VDR gene by radiation hybrid (RH) analysis, the isolation of a bacterial artificial chromosome (BAC) containing this gene, and physical mapping of the VDR gene by fluorescent in situ hybridization (FISH). RH analysis placed the VDR gene locus at chromosome 12cen‐q12, flanked by Stanford Human Genome Center (SHGC) 30216 and SHGC 9798 (D12S1892) markers. FISH analysis of a BAC containing the VDR gene confirmed its centromeric location. Thus, we have identified a BAC and genetic markers which can be used in the genetic analysis of the VDR gene and investigation of its involvement in osteoporosis and related disorders. We conclude that the VDR gene is centromeric to its previously reported locus on chromosome 12.
INTRODUCTION
The human vitamin D receptor gene (VDR) has been the object of intense investigation because of its potential involvement in the genetic determination of bone mineral density.(1) The VDR gene has been mapped to human chromosome 12 by somatic cell hybrid mapping,(2) in situ G‐banding,(3) and fluorescent in situ hybridization (FISH).(4,5) These studies concluded that VDR is located in the human 12q12‐q22 region,(2–5) in close proximity to the collagen type 2 α1 chain (COL2 A1) gene.(5) Until recently, the VDR gene's genomic structure was not known.(6) This hindered efforts to localize VDR relative to existing genetic markers(7) and physical contig maps(8) and to identify new polymorphic sequence tagged sites (STSs) for genetic studies of osteoporosis and related disorders.
In addition to polygenic osteoporosis, three monogenic endocrine disorders have been mapped to the most proximal (centromeric) human 12q area (listed elsewhere(9)). These are hypocalcemic vitamin D–resistant rickets (HVDRR), (Mendelian inheritance in man [MIM] catalog number 601769), pseudovitamin D–deficient rickets (PDDR) (MIM 264700), and triple A or Allgrove syndrome (AS) (MIM 231550), which are caused by mutations in the VDR gene, 25‐hydroxyvitamin D3–1α‐hydroxylase (P450c1α), and an as yet unidentified gene, respectively. As part of our linkage mapping studies in AS(10) and the genetic investigation of a family with HVDRR (Stratakis CA, Orban Z, and Barsony J, unpublished data), we sought to determine the precise genetic and physical location of the VDR gene on 12q. Meiotic mapping suggested that the location of the VDR gene is further centromeric than previously thought on chromosome 12. This was confirmed by radiation hybrid (RH) and bacterial artificial chromosome (BAC) FISH, both high‐resolution mapping techniques that have not previously been applied to the VDR gene.
MATERIALS AND METHODS
RH analysis
The 10,000 rad Stanford Human Genome Center (SHGC) G3 RH panel was used to determine the location of the VDR gene with regard to markers mapped to chromosome 12.(8,11) Polymerase chain reaction was performed with two sets of primers, one containing exon 4 (VDR‐4) and another containing exon 7 (VDR‐7) of the VDR gene.(6) These two amplicons were chosen because of the large intron separating exons 6 and 7 of the gene, ensuring that the position of the entire gene would be evaluated. The RH mapping data of both amplicons were submitted, without prior chromosomal assignment, to the Stanford Human Genome Center (SHGC) RH server to determine the most closely linked, previously mapped markers, as previously described.(8,11)
Identification of a BAC containing the VDR gene
BAC 325‐C‐22, containing both VDR‐4 and VDR‐7 amplicons of the VDR gene, was obtained from a commercially available library (Research Genetics, Birmingham, AL, U.S.A.), using the primer pairs mentioned above (data not shown). The BAC was grown overnight in 1 l LB‐chloramphenicol (12.5 μg/ml). The culture then was centrifuged, followed by DNA extraction. Extracted DNA was labeled for use in FISH.
Fluorescent in situ hybridization
FISH was performed following protocols described by Pinkel et al.(12) with minor modifications.(13) Briefly, metaphase spreads from peripheral leukocytes of a healthy donor were hybridized to digoxigenin‐11‐dUTP–labeled probe, followed by rhodamine‐conjugated antidigoxigenin antibodies (Boehringer‐Mannheim, Indianapolis, IN, U.S.A.). Hybridization and digital image analysis were performed as previously described.(14) Chromosomes were identified by converting 4′‐6‐diamidino‐2‐phenylindole (DAPI)‐banding into G‐simulated banding using the IP Lab Image software (Scan Analytics Corp., Vienna, VA, U.S.A.). Rehybridization with alpha‐satellite centromeric probes (Oncor, Gaithersburg, MD, U.S.A.) confirmed chromosomal localization.
RESULTS
VDR RH mapping and genetic localization
The VDR‐4 amplicon was indistinguishable from the chromosome 12 centromeric marker SHGC 9798 (also known as D12S1892), with a logarithm of odds (LOD) score of 1000 and an estimated distance of 0 cR10,000. The VDR‐7 amplicon mapped most closely to the chromosome 12 centromeric marker SHGC30216 (just centromeric to SHGC9798), with an LOD of 7.8 and an estimated distance of 8.5 cR10,000 (230 kb for chromosome 12).(11) The markers SHGC 9798 and SHGC 30216 are among the most centromeric framework markers on the long arm of chromosome 12, indicating that the VDR gene is centromeric to all of its previously reported locations in the proximal area of 12q.(2–5)
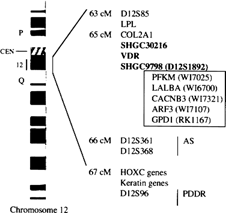
Based on these data, the currently available Whitehead Institute contig for human 12cen (WC12.1),(11) and other physical mapping information for the 12cen‐12q15 region,(9,15–18) the VDR gene is located ∼65 cM from the top of chromosome 12. This genetic locus contains several other genes and polymorphic STSs. These are listed as follows, with their database gene entries, corresponding STS, and distances from the top in parentheses, where applicable: 12cen‐D12S85 (63 cM); lipoprotein lipase (LPL, C114C11); D12S1661 (65 cM); vacuolar ATPase (WI‐8712); COL2 A1 (WI‐6991); VDR–muscle type phosphofructokinase (PFKM, WI‐7025); αLactalbumin (LALBA, WI‐7600); voltage‐dependent calcium channel β‐3 subunit (CACNB3, WI‐7321); ADP‐ribosylation factor 3 (ARF3, WI‐7107); glycerol‐3‐phosphate dehydrogenase (GPD1 [GPDH], RK11671168); D12S361 (66 cM); 12tel. This region corresponds to cytogenetic band 12cen‐q12. The neighboring homeobox C and keratin gene clusters flanked by genetic markers D12S368 and D12S96(10,18) correspond to cytogenetic band 12q13 and are also centromeric to the previously reported VDR locus at 12q14 (Fig. 1).
Ideogram of the long arm of chromosome 12, with G bands, and RH map of markers closely linked to VDR in 12cen‐q12. Markers SHGC30216 and SHGC9798 (the latter also known as D12S1892) flank the VDR gene at ∼65 cM from the top of human chromosome 12 (7, 8, 11). Other genes and disease loci in the region are also shown.
BAC containing the VDR gene and FISH analysis
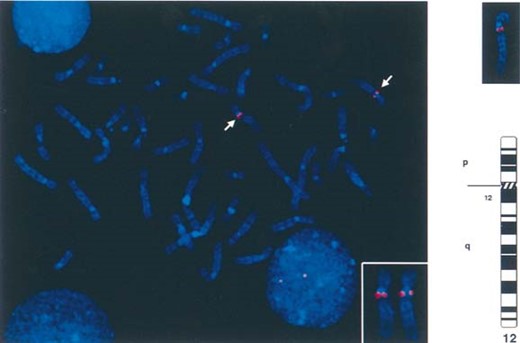
BAC 325‐C‐22, containing the VDR gene, was isolated and used as a FISH probe, along with the control centromeric chromosome 12 α‐satellite (Fig. 2). A total of 25 cells were examined. In all metaphases scored, a clear signal was seen on 12q very close to the centromere. DAPI banding unambiguously showed the position of the signal to be in the 12cen‐q12 region (Fig. 2).
DAPI‐counterstained chromosomes in metaphase showing the location of BAC 325‐C‐22 (which contains the VDR gene) on the most centromeric part of the long arm of chromosome 12. The two chromosome 12 homologs are identified by the positive α‐satellite signals (green) (see enclosed panel). In the right upper panel, examples of chromosome 12 color images which were converted to black and white “G‐band” images to show the relative position of the probe to the centromeric end of the long arm of chromosome 12. This region is identified as 12cen‐q12.
DISCUSSION
The VDR gene was the first among the steroid hormone receptor superfamily of proteins for which mutations were found to be responsible for human disease (HVDRR).(19) The subsequent association of osteoporosis with genetic polymorphisms in VDR gene introns(20) heightened the interest in this gene, which was genetically and physically mapped to chromosome 12.(2–5) However, the VDR gene, like many other genes, was originally assigned a location by somatic cell hybrid analysis and/or radioactive in situ hybridization.(2–5) Use of new physical and genetic mapping methods has shown that map assignments made with these methods may be misleading and should be considered to be approximations of the true locus.(21,22) In addition, new genomic methods can link VDR with STS that constitute the framework of the currently available maps of chromosome 12(7–9,11) and thus provide new genetic tools for the investigation of osteoporosis and related disorders.
Our data show that the VDR gene is located more centromerically than previously reported. RH placed VDR between markers SHGC 9798 (D2S1892) and SHGC 30216. VDR and SHGC 9798 were indistinguishable with the G3 RH panel, which has a resolution limit of 500 bp. The FISH data confirm the placement of VDR near the chromosome 12 centromere. This difference is explained by the expansion of the 12cen‐q15 gene map over the last few years. The 12cen‐q15 area appears to be especially gene‐rich, but is insufficiently covered by the existing contigs.(15) Therefore, the true location of other genes structurally or functionally related to VDR and preliminarily mapped to 12q13‐14 may also be more centromeric than previously reported. Examples include the COL2 A1 gene(17) and the genes encoding the transcription factor Sp‐1(23) and 25‐hydroxyvitamin D‐1α‐hydroxylase.(24) Inconsistencies in the association of VDR genotypes with osteoporosis(20,25) may also be explained when the physical map of the 12cen‐12q area is resolved. Of particular importance is the location of the COL2 A1 gene, which appears to be in close proximity to the VDR gene. It has been postulated that the association of VDR genotypes with knee osteoarthritis may be found because the VDR and COL2 A1 genes are in linkage disequilibrium.(26) Although this might be true, the distance between the two genes is unknown. It is possible that this genomic area may in fact be larger than expected from the genetic studies, because Col2a1 maps to mouse chromosome 15, an area syntenic to human 12qcen‐14, whereas vdr maps to mouse chromosome 5, an area syntenic to human 12q24.(27)
We conclude that the human VDR gene maps to 12cen‐q12. For genetic studies of disorders of the bone and cartilage, it appears that the area of human chromosome 12 contained between polymorphic markers D12S85 and D12S361 harbors both the COL2 A1 and VDR genes, although their precise order and physical distance remain unclear.
REFERENCES
Mouse Genome Database (MGD)