Abstract
The prion protein gene (PRNP) plays a central role in the origin of Creutzfeldt–Jakob disease (CJD), but there is growing interest in other polymorphisms that may be involved in CJD. Polymorphisms upstream of PRNP that may modulate the prion protein production as well as polymorphisms in the prion-like doppel gene (PRND) have been studied, with inconsistent findings. We investigated the role of a single-nucleotide polymorphism (SNP 1368) located upstream of PRNP and three polymorphisms in PRND (T26M, P56L and T174M) in CJD. The study included a population-based sample of 52 patients with sporadic CJD and 250 controls. We analysed our data as single markers and haplotypes. Further, we conducted a meta-analysis on PRND T174M comparing the data of the four studies conducted to date. For SNP 1368 and PRNP M129V, we found significant evidence for linkage disequilibrium. No evidence was found for a relation of SNP 1368 to CJD independent of PRNP M129V. We further found a significant increased prevalence of M homozygotes at PRND T174M among sporadic CJD patients, when adjusting the analyses for the other genotypes. In the haplotype analyses, the association was strongest for persons homozygous for PRNP 129M and PRND 174M (odds ratio 4.35, 95% confidence interval 1.05–8.09; P=0.04). The meta-analysis on the PRND T174M polymorphism did not show a consistent effect across studies, raising the question as to whether PRND 174M is causally related to CJD, or whether the PRND allele is in linkage disequilibrium with another polymorphism related to CJD.
Similar content being viewed by others
Introduction
The prion protein gene (PRNP) plays a central role in the origin of Creutzfeldt–Jakob disease (CJD). Susceptibility for the disease is influenced by a polymorphism at codon 129 of PRNP.1 At this site, homozygosity for methionine increases the risk for sporadic, iatrogenic and variant CJD, but also those homozygous for valine at codon 129 were found to be at increased risk for CJD.2,3,4 In recent years, there has been growing interest in other genetic polymorphisms involved in CJD. Since animal studies have shown that higher levels of PRNP expression lead to shorter incubation times,5,6 polymorphisms in the regulatory region of PRNP may be of interest. Several polymorphisms were identified in intronic and upstream regions of human PRNP.7,8 A single-nucleotide polymorphism (SNP) upstream PRNP exon 1, designated SNP 1368, was found to be strongly associated with CJD.7
Another gene of interest is the downstream prion protein-like gene, or doppel gene (PRND). This gene shows a 24% coding sequence identity with PRNP and is located 27 kb downstream of PRNP.9 The function of both proteins is still unresolved. Possibly, PrP and Dpl have antagonistic biological properties in neurons, for example, competitive binding to a common receptor. Deregulation of this balance may lead to neuropathology.9,10 In humans, several polymorphisms have been described in PRND. However, findings on the association of these polymorphisms with CJD are inconsistent.11,12,13
In order to study the effect of these potential susceptibility genes for CJD, we have examined the association of a polymorphism upstream PRNP and three polymorphisms in PRND, with sporadic CJD (Figure 1). We performed this association study in a population-based sample of 52 patients with sporadic CJD and 250 controls. The data on PRND T174M were analysed further in a meta-analysis, including all studies in sporadic CJD published to date.
Material and methods
Study population
CJD patients were derived from a population-based study in the Netherlands, which aimed to ascertain all patients with this disease. The patients in this study (n=52) were diagnosed with the sporadic form of CJD and were classified as definite or probable according to established criteria.14,15 The mean age of the patients was 63 years (range 30–82 years) and 63% were female. A medical doctor from the reference centre visited the patients and blood was drawn for DNA collection after informed consent of a relative. CJD patients were compared with a control group, free of dementia and randomly drawn from a population-based study, the Rotterdam Study, for which all inhabitants of a suburb of Rotterdam were invited to participate.16 The mean age of the control group was 67 years (range 55–87 years) and 57% were female.
Laboratory analyses
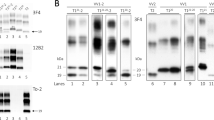
Genomic DNA was extracted from leucocytes following standard procedures.17 PRNP M129V genotype was determined as described in detail elsewhere.18 Briefly, DNA was amplified by polymerase chain reaction (PCR) producing a 722-bp fragment. A second, ‘nested’ PCR reaction produced a 95-bp fragment. The restriction enzyme BbrPI digested the PCR fragment producing a 75-bp fragment in case of codon 129V and 95-bp fragment in case of a codon 129M allele. The products were fractionated on a 3.5% agarose gel stained with ethidium bromide.
PRND T174M: Amplification of a 141-bp product in the PRND open-reading frame was performed with the forward primer 5′-CTC TGC CTT CTG GCT TTG AT-3′ and the reverse primer 5′-CTG GGC ACC TTC AGA ACA C-3′. The PCR product was digested by the restriction enzyme NlaIII into two fragments of 111 and 30 bp, when the M allele was present. The restriction product was fractionated on a 3.0% agarose gel stained with ethidium bromide.
PRND T26M, P56L and PRNP SNP 1368: The 528-bp coding sequence of PRND was amplified by PCR using the forward primer 5′-GAC CCA CCG CCG TTT CTCT-3′ and the reverse primer 5′-CGT GGG TTT GGG GGA GAA CA-3′ in standard conditions.11 For PCR amplification of SNP 1368, we designed the forward primer 5′-AAC CAG AAA CAT GGG GTG TT-3′ and the reverse primer 5′-AAA CCC TGG CTT TTA CAA AGA A-3′, producing a 167-bp fragment. The PCR products were purified with Sap 1 U/μl and Exo1 10 U/μl. A multiplex SNaPshot reaction (Applied Biosystems) was performed with the single base extension (SBE) primer for T26M 5′-CCA CCT CTC TGC GGT CCA GA-3′ and the primer for P56L 5′-polyT CCA GGT GGC TGA GAA CCG CC-3′. For SNP 1368, we used the primer 5′-polyT TGT TAA ATC AAT TAC AGG AG-3′. The SNaPshot products were electrophoresed on an ABI 3100 automated DNA sequencer (16, filter E, LIZ 120 as size standard, from Applied Biosystems). All SNPs analyses were performed with Genotyper Version 3.7.
Statistical analyses
Genotype and allele frequencies were tested for Hardy–Weinberg equilibrium (HWE) proportions.19 First, we tested for the association of the individual polymorphisms with CJD. We used logistic regression analysis to estimate odds ratios (OR) and their corresponding 95% confidence intervals (CI), while adjusting for the other polymorphisms. Based on the findings of previous studies assessing the risk of PRNP M129V, we selected the heterozygotes as the reference group, as this genotype is consistently associated with the lowest risk of CJD.2 For the other polymorphisms, we considered the most frequent allele in controls as the reference. Next, we assessed whether linkage disequilibrium existed between the polymorphisms using a generalised linear mixed model with random effects. Since parental information was missing, an expectation maximisation (E–M) algorithm was used to estimate haplotype frequencies in a multiple locus system in the samples.20 Finally, we studied the association of CJD with the haplotypes.
Meta-analysis
For the meta-analysis on the association between sporadic CJD and the PRND T174M polymorphism four studies (including this study) were available, leading to a total of 239 patients and 584 controls.11,12,13 Genotype frequencies were not in HWE proportions in the controls in one study (P=0.002).13 The meta-analysis was performed including and excluding this study, using the homozygotes for the most common allele as reference. Following the hypothesis that the prion protein and doppel protein may be involved in CJD according to a similar genetic mechanism, we also analysed homozygotes versus heterozygotes. The meta-analysis was not adjusted for the PRNP M129V polymorphism. Heterogeneity in allele frequencies between the studies was tested with a χ2 test. We used a random effect model when significant evidence for between study variation was found.21 In all other comparisons we used a fixed effect model.
Results
All genotype frequencies were in HWE proportions, with the exception of the PRNP M129V polymorphism in CJD patients. In CJD patients, those homozygous for the PRNP M129V polymorphism were over-represented (unadjusted OR 2.20, 95% CI 1.07–4.92; P=0.03). PRND T26M and P56L were found to be rare polymorphisms (Table 1), which were not associated with the disease. In Table 2, the allele and genotype distributions of SNP 1368, PRNP M129V and PRND T174M are shown. After adjusting for the other genotypes, we found a more than three times increased risk of CJD for carriers of PRNP 129 VV genotype compared with heterozygotes (OR 3.22, 95% CI 1.00–10.45; P=0.05). Those homozygous for MM at PRNP 129 were also at increased risk, although not significantly. A similar increase in risk of CJD to that seen for PRNP 129V homozygotes was found for MM carriers of PRND 174, compared to the reference group TT (OR 2.86, 95% CI 1.10–7.48; P=0.03). We found no association with CJD for SNP 1368.
We found a significant evidence for linkage disequilibrium between PRNP SNP 1368 and PRNP M129V, both in cases (P=0.0003) and controls (P=0.0001), with an over-representation of the combination SNP 1368t with codon 129V and SNP 1368c with codon 129M. However, the distribution of haplotypes did not differ significantly between cases and controls (P=0.24). In the analyses stratified on PRNP codon 129, we found the c-allele slightly over-represented in homozygous carriers of PRNP codon 129M, although this finding was not statistically significant. We found no evidence for linkage disequilibrium between PRNP M129V and PRND T174M (P=0.31 in cases and P=0.17 in controls). We observed a significant association between haplotype PRNP codon 129M–PRND codon 174M and sporadic CJD (OR 1.88, 95% CI 1.41–3.28). Also, the frequency of the PRNP codon 129V–PRND codon 174M haplotype was increased in sporadic CJD, but not significantly (OR 1.81, 95% CI 0.91–4.08).
In Table 3, the haplotype interaction is assessed. In the haplotypes, the first locus is the PRNP M129V polymorphism and the second locus the PRND T174M polymorphism. We used the MT-VT genotype as the reference group, because in PRND T174M we found the M allele to be the risk allele and T the wild-type allele. Among sporadic CJD patients, there was a significant increase in carriers of MM-MM (OR 4.35, 95% CI 1.05–18.09; P=0.04). Although the frequencies of other haplotypes was increased, numbers were too small to reach statistical significance.
The meta-analysis on the studies assessing the relation between the PRND T174M polymorphism and CJD did not result in significant findings (Figure 2), both in the analyses including and excluding the study of Schröder et al.13 P-values for the overall effect of the T174M genotype on CJD ranged from 0.4 to 1.0. Also, when we tested homozygosity versus heterozygosity, we did not observe an effect of the T174M polymorphism on CJD, neither in the analysis including nor excluding the study of Schröder et al (data not shown).
Discussion
We found a significant relation of PRNP M129V and PRND T174M with the sporadic form of CJD. Consistent with the existing literature, we found homozygosity for PRNP 129 associated with an increased risk for the sporadic form of CJD.1 The PRND T174M polymorphism was significantly associated with CJD, independent of the effect of the PRNP M129V polymorphism, resulting in an almost three-fold increased risk for M homozygotes. This finding is not in line with earlier reports, which either showed no significant relation11,12 or a significant increased risk for the heterozygotes.13 A problem with the latter study is that the controls were not in HWE (P=0.002). This hampers the interpretation of the study since deviations from HWE may be the result of selection bias or genotyping errors.
Two other studies assessing the relation of the upstream region of human PRNP with CJD have been conducted.7,8 In one of these studies, the effect of SNP 1368 was assessed, which was found to be related to CJD.7 This study found strong linkage disequilibrium between SNP 1368 and PRNP codon 129, which was confirmed in our study. In both studies, the SNP 1368c allele was found most frequently in combination with PRNP 129M.7 In our study, the risk associated with the SNP 1368c allele was only slightly elevated and the association between PRNP SNP 1368 and sporadic CJD was not statistically significant. This may be explained by differences across the studies in linkage disequilibrium for SNP 1368 and a functional polymorphism nearby. In Caucasian populations, linkage disequilibrium with polymorphisms upstream the gene may differ considerably.22
A problem in the interpretation of our data is the small number of patients. In order to increase the power of our study, we included five controls per case. Further, we matched the controls on age, to avoid spurious associations based on survival of one of the genotypes. Despite the lack of power, our study suggests a significant and consistent effect of PRNP T174M. The strength of our study is its population-based design in both the case and control series. Although the distributions of the genotypes in the studies are similar,7,11,12 the meta-analysis suggests significant differences in the effect of the genotypes across studies, also when the study in which the controls were not in HWE was excluded.13 There may be various explanations for these differences. The small size of each of the studies may have resulted in random fluctuations. Another explanation is that the PRND T174M is not the causal locus, but may be in linkage disequilibrium with a nearby locus involved in CJD. Linkage disequilibrium may differ across populations and this may explain the differences observed between the studies. Further crosscultural studies are needed to resolve this issue.
Although we cannot pinpoint the causal variant, the findings in CJD patients are compatible with a role of Dpl in the sporadic form of CJD. The function of Dpl remains uncertain, but may overlap the function of PrP. Overexpression of Dpl in PrP knockout mice with major deletions in PRNP has been shown to result in progressive ataxia.23 Recent evidence suggests that truncated PrP and Dpl may interfere with a signalling pathway essential for the maintenance of cell viability.24 However, it is unclear whether these findings in mice reflect the situation in humans. In CJD patients and controls, no difference in brain Dpl levels was observed, suggesting that Dpl levels may not be influenced by prion infection.25
In conclusion, our study suggests that the T174M polymorphism in PRND may increase the risk for CJD. However, given the small size of our study and those of others, further research on the role of PRND in the aetiology of CJD is needed.
References
Palmer MS, Dryden AJ, Hughes JT, Collinge J : Homozygous prion protein genotype predisposes to sporadic Creutzfeldt–Jakob disease. Nature 1991; 352: 340–342.
Alperovitch A, Zerr I, Pocchiari M et al: Codon 129 prion protein genotype and sporadic Creutzfeldt–Jakob disease. Lancet 1999; 353: 1673–1674.
Collinge J, Palmer MS, Dryden AJ : Genetic predisposition to iatrogenic Creutzfeldt–Jakob disease. Lancet 1991; 337: 1441–1442.
Kovacs GG, Head MW, Bunn T et al: Clinicopathological phenotype of codon 129 valine homozygote sporadic Creutzfeldt–Jakob disease. Neuropathol Appl Neurobiol 2000; 26: 463–472.
Westaway D, Mirenda CA, Foster D et al: Paradoxical shortening of scrapie incubation times by expression of prion protein transgenes derived from long incubation period mice. Neuron 1991; 7: 59–68.
Collinge J, Palmer MS, Sidle KC et al: Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature 1995; 378: 779–783.
Mead S, Mahal SP, Beck J et al: Sporadic – but not variant – Creutzfeldt–Jakob disease is associated with polymorphisms upstream of PRNP exon 1. Am J Hum Genet 2001; 69: 1225–1235.
McCormack JE, Baybutt HN, Everington D et al: PRNP contains both intronic and upstream regulatory regions that may influence susceptibility to Creutzfeldt–Jakob disease. Gene 2002; 288: 139–146.
Mastrangelo P, Westaway D : The prion gene complex encoding PrP(C) and doppel: insights from mutational analysis. Gene 2001; 275: 1–18.
Cui T, Holme A, Sassoon J, Brown DR : Analysis of doppel protein toxicity. Mol Cell Neurosci 2003; 23: 144–155.
Peoc'h K, Guerin C, Brandel JP et al: First report of polymorphisms in the prion-like protein gene (PRND): implications for human prion diseases. Neurosci Lett 2000; 286: 144–148.
Mead S, Beck J, Dickinson A et al: Examination of the human prion protein-like gene doppel for genetic susceptibility to sporadic and variant Creutzfeldt–Jakob disease. Neurosci Lett 2000; 290: 117–120.
Schröder B, Franz B, Hempfling P et al: Polymorphisms within the prion-like protein gene (Prnd) and their implications in human prion diseases, Alzheimer's disease and other neurological disorders. Hum Genet 2001; 109: 319–325.
Masters CL, Harris JO, Gajdusek DC et al: Creutzfeldt–Jakob disease: patterns of worldwide occurrence and the significance of familial and sporadic clustering. Ann Neurol 1979; 5: 177–188.
Poser S, Mollenhauer B, Kraubeta A et al: How to improve the clinical diagnosis of Creutzfeldt–Jakob disease. Brain 1999; 122: 2345–2351.
Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA : Determinants of disease and disability in the elderly: the Rotterdam elderly study. Eur J Epidemiol 1991; 7: 403–422.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Van Everbroeck B, Croes EA, Pals P et al: Influence of the prion protein and the apolipoprotein E genotype on the Creutzfeldt–Jakob disease phenotype. Neurosci Lett 2001; 313: 69–72.
Terwilliger JD OJ: Handbook of Human Genetic Linkage. New York: John Hopkins University Press; 1994.
Chiano MN, Clayton DG : Fine genetic mapping using haplotype analysis and the missing data problem. Ann Hum Genet 1998; 62: 55–60.
Normand SL : Meta-analysis: formulating, evaluating, combining, and reporting. Stat Med 1999; 18: 321–359.
Roks G, Cruts M, Bullido MJ et al: The −491 A/T polymorphism in the regulatory region of the apolipoprotein E gene and early-onset Alzheimer's disease. Neurosci Lett 1998; 258: 65–68.
Moore RC, Lee IY, Silverman GL et al: Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J Mol Biol 1999; 292: 797–817.
Flechsig E, Hegyi I, Leimeroth R et al: Expression of truncated PrP targeted to Purkinje cells of PrP knockout mice causes Purkinje cell death and ataxia. EMBO J 2003; 22: 3095–3101.
Peoc'h K, Volland H, De Gassart A et al: Prion-like protein doppel expression is not modified in scrapie-infected cells and in the brains of patients with Creutzfeldt–Jakob disease. FEBS Lett 2003; 536: 61–65.
Acknowledgements
This study would not have been possible without the cooperation of all Dutch neurologists and the kind help of the family members of the patients. This work was supported by the Dutch Ministry of Health, Welfare and Sports (VWS), an EU grant (CT98-7022) and the Netherlands Organisation for Scientific Research (NWO); the Fund for Scientific Research Flanders (FWO-F), DWTC Interuniversity Attractionpoles (IUAP), the International Alzheimer's Research Foundation (IARF), the Alzheimer Association (AA) and the University of Antwerp (UIA), Belgium. Bart Dermaut is a PhD fellow of the FWO-F.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Croes, E., Alizadeh, B., Bertoli-Avella, A. et al. Polymorphisms in the prion protein gene and in the doppel gene increase susceptibility for Creutzfeldt–Jakob disease. Eur J Hum Genet 12, 389–394 (2004). https://doi.org/10.1038/sj.ejhg.5201161
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201161
Keywords
This article is cited by
-
RARB and STMN2 polymorphisms are not associated with sporadic Creutzfeldt–Jakob disease (CJD) in the Korean population
Molecular Biology Reports (2014)
-
A polymorphism in the YWHAH gene encoding 14-3-3 eta that is not associated with sporadic Creutzfeldt–Jakob disease (CJD)
Molecular Biology Reports (2012)
-
Analysis of single nucleotide polymorphisms of PRNP gene in twenty-four ethnic groups of India
Journal of Genetics (2010)
-
Lack of association between PRNP 1368 polymorphism and Alzheimer's disease or vascular dementia
BMC Medical Genetics (2009)
-
A critical analysis of disease-associated DNA polymorphisms in the genes of cattle, goat, sheep, and pig
Mammalian Genome (2008)