Abstract
Cardiofaciocutaneous (CFC) syndrome is a multiple congenital anomalies/mental retardation syndrome characterized by congenital heart defects, characteristic facial appearance, short stature, ectodermal abnormalities and mental retardation. It was described in 1986, and to date is of unknown genetic etiology. All reported cases are sporadic, born to non-consanguineous parents and have apparently normal chromosomes. Noonan and Costello syndromes remain its main differential diagnosis. The recent finding of PTPN11 missense mutations in 45–50% of the Noonan patients studied with penetrance of almost 100% and the fact that in animals mutations of this gene cause defects of semilunar valvulogenesis, made PTPN11 mutation screening in CFC patients a matter of interest. We sequenced the entire coding region of the PTPN11 gene in ten well-characterised CFC patients and found no base changes. We also studied PTPN11 cDNA in our patients and demonstrated that there are no interstitial deletions either. The genetic cause of CFC syndrome remains unknown, and PTPN11 can be reasonably excluded as a candidate gene for the CFC syndrome, which we regard as molecular evidence that CFC and Noonan syndromes are distinct genetic entities.
Similar content being viewed by others
Introduction
Cardiofaciocutaneous (CFC) syndrome (MIM 115150) is a multiple congenital anomalies/mental retardation syndrome characterised by congenital heart defects, characteristic facial appearance, short stature, ectodermal abnormalities, and mental retardation.1 A recent review has identified 54 published classical cases of CFC syndrome. All cases were sporadic, with apparently normal chromosomes, born to non-consanguineous parents and presenting the same unique combination of findings and natural history.2
The CFC phenotype comprises a characteristic facial appearance, congenital heart defects (pulmonic stenosis, 37%; atrial septal defect, 35%; hypertrophic cardiomyopathy, 11%), mental retardation/developmental delay (81%), speech delay (46%), short stature (78%) and hypotonia (28%). The typical craniofacial manifestations include relative macrocephaly, high forehead, bitemporal constriction, supraorbital hypoplasia, short nose with depressed nasal bridge and anteverted nostrils, apparently low-set and posteriorly angulated ears, hypertelorism, ptosis, epicanthal folds, and downslanting of palpebral fissures. All patients had an ectodermal abnormality consisting of sparse, slowly growing and curly hair, sparse or absent eyelashes and eyebrows, follicular keratosis or ulerythema ophryogenes, ichthyosis, hyperkeratosis, generalized hyperpigmentation, haemangiomas, hyperelastic skin, cutis marmorata, café-au-lait patches, and/or slowly growing nails.
In spite of the controversy about the distinction between CFC and Noonan syndrome (NS) most of the time a refined clinical evaluation is sufficient for the differential diagnosis. Although phenotypic similarity between CFC and NS exists, especially regarding cardiac defects, craniofacial findings, short stature and congenital lymphedema, there are clear differences in clinical manifestations and natural history that separate the two conditions.3,4,5
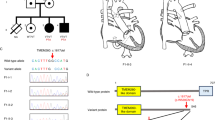
Autosomal dominant inheritance is evident in many NS families, and genetic heterogeneity has also been demonstrated.6 Linkage studies have defined a region of approximately 7.5 Mb on the long arm of chromosome 12 (12q24) as a candidate region for a gene(s) causing NS6,7,8,9 (Figure 1). The recent demonstration of mutations in PTPN11, the gene encoding the non-receptor-type protein tyrosine phosphatase SHP-2 (src homology region 2-domain phosphatase-2)10 (Figure 2), in 45–50% of the studied patients of NS,9,11 has enabled an initial genotype–phenotype analysis. This has also shown that pulmonic stenosis is more prevalent in the group of NS patients who had a PTPN11 mutation than in the group without a mutation (70.6 vs 46.2%; P<0.01). The identification of a PTPN11 mutation in a family segregating the Noonan-like/multiple giant-cell lesion syndrome,11 and in LEOPARD syndrome,12 suggests a considerable phenotypic range due to mutations in this gene making it necessary to study the possible role of this gene in CFC syndrome.
Patients
We studied ten CFC patients (nine males and one female) (Figure 3), randomly selected from a larger group of CFC patients followed by a group of clinical geneticists in Brazil, Italy and North America, experienced with CFC syndrome and its nosology with the Noonan and Costello syndromes. In all cases the clinical diagnosis was considered incontrovertible. The CFC Index2 was applied to all patients. The patients selected for this study are originally from Brazil and Italy and in each case a consent form was signed by the parents.
Materials and methods
DNA extraction
Genomic DNA was isolated from the patients' peripheral-blood leukocytes or from Epstein–Barr virus-transformed lymphocytes using standard procedures.
Gene sequencing
The 15 exons of the PTPN11 gene, described at http:// genome.ucsc.edu//cgi-bin/hgTracks?position=NM 002834& Submit=Submit and at http://www.ncbi.nlm.nih.gov/entrez / viewer.fcgi?dispmax =1&showndispmax = 1&val = NT 009575.8&view=graph& from=758671& to=857626& sfrom=0& strand=2, and their intron–exon boundaries were sequenced bi-directionally (Table 1, Figure 2). These regions were amplified from genomic DNA using the primers shown in Table 2, which were selected by the Macintosh program Primer Express.
PRC reactions were carried out under standard conditions, containing: 50 ng of genomic DNA, 1 mM of each forward and reverse primers and 10 μl of Promega TAX Master Mix, as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C (1 min), annealing at 53–60°C (1 min) and extension at 72°C (1 min), followed by final extension at 72°C for 5 min, using the Gene Amp PCR System 9600 - Perkin Elmer.
PRC products were purified with a PCR product pre-sequencing kit, composed of exonuclease 1 and shrimp alkaline phosphatase, by USB Corporation, according to their instructions, and later using the Centri-Sep Spin Columns by Applied Biosystems. Sequencing reactions were carried out using the ABIPRISM dye terminator cycle sequencing kit, and the products were analysed on an ABIPRISM 310 Genetic Analyzer DNA sequencer. Sequences were analysed manually.
Study of the cDNA
In order to detect the presence of deletions within the coding region, which could be overlooked by analysing the sequences, we constructed five pairs of primers to amplify overlapping cDNA fragments of about 550 bp (Table 3). These fragments were amplified by PCR, carried out under standard conditions, containing: 50 ng of cDNA, 1 mM of each forward and reverse primers and 10 μl of Promega TAX Master Mix, as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C (1 min), annealing at 55–64°C (1 min) and extension at 72°C (1.5 min), followed by extension period of 10 min, using the Gene Amp PCR System 9600 - Perkin Elmer. Every PCR reaction included 10 CFC patients and one control subject. PCR products were electrophoresed on a 2% agarose gel to verify the actual size of the fragments obtained from each patient.
Results
The CFC Index of all ten patents included in this study ranged from −1 SD to +1 SD. Three facial pictures are shown in Figure 3.
The coding-region of PTPN11 and its intron–exon boundary regions were sequenced in 10 CFC patients and no base changes were identified.
The sizes of the amplified cDNA fragments were as expected and single bands were detected from all patients in all five fragments.
Discussion
The actual relationship between CFC and Noonan syndromes has been the object of a heated controversy, and many opinions have been expressed as to whether they are different genetic entities, allelic conditions, a contiguous gene syndrome, or even different phenotypes of the same condition.2,3,4,5,13,14,15 The finding of one of the causes of Noonan syndrome is a very important step towards the clarification of this matter.
The protein encoded by PTPN11 is a member of the protein tyrosine phosphatase (PTP) family. PTPs regulate a variety of cellular processes, including cell growth, differentiation, mitotic cycle and oncogenic transformation and, in mouse, is involved in cardiac semilunar valvogenesis.16 The finding of 45–50% PTPN11 mutation prevalence in a sample of Noonan syndrome patients with almost 100% penetrance9,11 demonstrates that it is one of the genes causing Noonan syndrome. Mutations found in NS patients were all missense changes affecting amino acid residues that are conserved among the vertebrate SHP-2 orthologs.
PTPN11 mutations have also been found in conditions such as the Multiple-Lentigines/LEOPARD syndrome12 and Noonan-like/multiple giant-cell lesion syndrome.11 These data are of great importance in extending the phenotype of Noonan syndrome. It is also important to better classify some patients whose diagnosis is controversial. The finding of the same PTPN11 mutation in the two patients from a large family with NS who were previously diagnosed as having CFC syndrome17,18 demonstrates that those patients also have Noonan syndrome as already evident by their published phenotype. Therefore, to date, there are no familial cases of CFC syndrome. We have demonstrated that CFC syndrome patients do not have missense changes on the coding region of the PTPN11, nor small or large intragenic deletions, which we regard as molecular evidence that CFC and Noonan syndromes are distinct genetic entities.
We encourage PTPN11 mutation screening in other CFC patients to confirm our results.
References
Reynolds JF, Neri G & Herrmann JP et al: New multiple congenital anomalies/mental retardation syndrome with cardio-facio-cutaneous involvement – the CFC syndrome. Am J Med Genet 1986; 25: 413–427.
Kavamura MI, Peres CA, Alchorne MMA & Brunoni D : CFC Index for the diagnosis of cardiofaciocutaneous syndrome. Am J Med Genet 2002; 112: 12–16.
Neri G, Zollino M & Reynolds JF : The Noonan–CFC controversy. Am J Med Genet 1991; 39: 367–370, [editorial comment]
Neri G & Zollino M : More on the Noonan-CFC controversy. Am J Med Genet 1996; 65: 100, [editorial comment]
Grebe TA & Clericuzio C : Neurologic and gastrointestinal dysfunction in cardio-facio-cutaneous syndrome: identification of a severe phenotype. Am J Med Genet 2000; 95: 135–143.
Jamieson CR, Van Der Brugt I & Brady AF et al: Mapping a gene for Noonan syndrome to the long arm of chromosome 12. Nat Genet 1994; 8: 357–360.
Legius E, Schollen E, Matthijs G & Fryns JP : Fine mapping of Noonan/cardio-facio-cutaneous syndrome in a large family. Eur J Hum Genet 1998; 6: 32–37.
Ogata T, Muroya K & Tsukahara M : Noonan syndrome: genotype analysis of the Noonan syndrome critical region at chromosome 12q in a three-generation family. Am J Med Genet 1998; 79: 153–154.
Tartaglia M, Mehler EL & Goldberg R et al: Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 2001; 29: 465–468.
Hof P, Pluskey S, Dhe-Paganon S, Ect MJ & Shoelson SE : Crystal structure of the tyrosine phosphatase SHP-2. Cell 1998; 92: 441–450.
Tartaglia M, Kalidas K & Shaw A et al: PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet 2002; 70: 1555–1563.
Digilio MC, Conti E & Sarkozy A et al: Grouping of Multiple-Lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet 2002; 71: 389–394.
Fryer AE, Holt PJ & Hughes HE : The Cardio-facio-cutaneous (CFC) syndrome and Noonan syndrome: Are they the same?. Am J Med Genet 1991; 38: 548–551.
Leichtman LG : Are cardio-facio-cutaneous syndrome and Noonan syndrome distinct? A case of CFC offspring of a mother with Noonan syndrome. Clin Dysmorph 1996; 5: 61–64.
Lorenzetti ME & Fryns JP : Retinitis pigmentosa in a young man with Noonan syndrome: Further evidence that Noonan syndrome (NS) and the cardio-facio-cutaneous syndrome (CFC) are variable manifestations of the same entity?. Am J Med Genet 1996; 65: 97–99.
Chen B, Bronson RT & Klaman LD et al: Mice mutants for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet 2000; 24: 296–299.
Fryns JP, Volcke PH & Van Den Berghe H : The Cardio-facio-cutaneous (CFC) syndrome: autosomal dominant inheritance in a large family. Genet Counsel 1991; 3: 19–24.
Schollen E, Matthijs G, Legius E & Fryns J : Mutation in the gene for protein tyrosine phosphatase SHP-2 (PTPN11) in a large family with Noonan/cardio-facio-cutaneous syndrome. Eur J Hum Genet 2002; 10, (Suppl 1): 238, [abstract].
Acknowledgements
We thank the CFC Family Network and its President, Brenda Conger and CAPES-FADA (Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), São Paulo - SP, Brazil, for financial support and for their contribution to this work. We also thank Drs: Giuseppina Corona, Livia Garavelli, Fernanda Teresa de Lima, Debora Pallos, Maria Piccione, Jane Sanchez and Gioacchino Scarano, for referral of patients. We thank Drs Charles Schwartz and Ilaria Zito for their valuable suggestions to this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kavamura, M., Pomponi, M., Zollino, M. et al. PTPN11 mutations are not responsible for the Cardiofaciocutaneous (CFC) syndrome. Eur J Hum Genet 11, 64–68 (2003). https://doi.org/10.1038/sj.ejhg.5200911
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200911
Keywords
This article is cited by
-
What’s new in the neuro-cardio-facial-cutaneous syndromes?
European Journal of Pediatrics (2007)
-
Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome
Nature Genetics (2006)
-
Noonan syndrome and related disorders: Alterations in growth and puberty
Reviews in Endocrine and Metabolic Disorders (2006)