Abstract
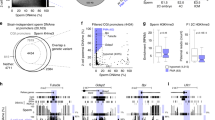
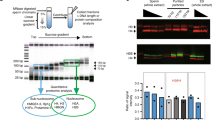
In higher eukaryotes, histone methylation is involved in maintaining cellular identity during somatic development. As most nucleosomes are replaced by protamines during spermatogenesis, it is unclear whether histone modifications function in paternal transmission of epigenetic information. Here we show that two modifications important for Trithorax- and Polycomb-mediated gene regulation have methylation-specific distributions at regulatory regions in human spermatozoa. Histone H3 Lys4 dimethylation (H3K4me2) marks genes that are relevant in spermatogenesis and cellular homeostasis. In contrast, histone H3 Lys27 trimethylation (H3K27me3) marks developmental regulators in sperm, as in somatic cells. However, nucleosomes are only moderately retained at regulatory regions in human sperm. Nonetheless, genes with extensive H3K27me3 coverage around transcriptional start sites in particular tend not to be expressed during male and female gametogenesis or in preimplantation embryos. Promoters of orthologous genes are similarly modified in mouse spermatozoa. These data are compatible with a role for Polycomb in repressing somatic determinants across generations, potentially in a variegating manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Chong, S. & Whitelaw, E. Epigenetic germline inheritance. Curr. Opin. Genet. Dev. 14, 692–696 (2004).
Hochedlinger, K. & Jaenisch, R. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N. Engl. J. Med. 349, 275–286 (2003).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Puschendorf, M. et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 (2008).
Albert, M. & Peters, A.H. Genetic and epigenetic control of early mouse development. Curr. Opin. Genet. Dev. 19, 113–121 (2009).
Balhorn, R., Gledhill, B.L. & Wyrobek, A.J. Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry 16, 4074–4080 (1977).
Gatewood, J.M., Cook, G.R., Balhorn, R., Bradbury, E.M. & Schmid, C.W. Sequence-specific packaging of DNA in human sperm chromatin. Science 236, 962–964 (1987).
Gardiner-Garden, M., Ballesteros, M., Gordon, M. & Tam, P.P. Histone- and protamine-DNA association: conservation of different patterns within the β-globin domain in human sperm. Mol. Cell. Biol. 18, 3350–3356 (1998).
Wykes, S.M. & Krawetz, S.A. The structural organization of sperm chromatin. J. Biol. Chem. 278, 29471–29477 (2003).
Pittoggi, C. et al. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J. Cell Sci. 112, 3537–3548 (1999).
Hammoud, S.S. et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009).
van der Heijden, G.W. et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev. Biol. 8, 34 (2008).
Sparmann, A. & van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856 (2006).
Hublitz, P., Albert, M. & Peters, A.H. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 53, 335–354 (2009).
Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002).
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 (2002).
Shen, X. et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008).
Mohn, F. et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008).
Ezhkova, E. et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135 (2009).
Margueron, R. et al. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Hansen, K.H. et al. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008).
Alsheimer, M., Fecher, E. & Benavente, R. Nuclear envelope remodelling during rat spermiogenesis: distribution and expression pattern of LAP2/thymopoietins. J. Cell Sci. 111, 2227–2234 (1998).
Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Weber, M. et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39, 457–466 (2007).
Imamura, M. et al. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 6, 34 (2006).
Pan, G. et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299–312 (2007).
Zhao, X.D. et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286–298 (2007).
Bracken, A.P., Dietrich, N., Pasini, D., Hansen, K.H. & Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006).
Chalmel, F. et al. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA 104, 8346–8351 (2007).
Geremia, R., Boitani, C., Conti, M. & Monesi, V. RNA synthesis in spermatocytes and spermatids and preservation of meiotic RNA during spermiogenesis in the mouse. Cell Differ. 5, 343–355 (1977).
Namekawa, S.H. et al. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16, 660–667 (2006).
Zeng, F. & Schultz, R.M. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283, 40–57 (2005).
Ohta, H., Tohda, A. & Nishimune, Y. Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol. Reprod. 69, 1815–1821 (2003).
Anderson, E.L. et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA 105, 14976–14980 (2008).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Strumpf, D. et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 (2005).
Ng, R.K. et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 (2008).
Pan, H., O'Brien, M.J., Wigglesworth, K., Eppig, J.J. & Schultz, R.M. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev. Biol. 286, 493–506 (2005).
Faulkner, G.J. et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41, 563–571 (2009).
Arpanahi, A. et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 19, 1338–1349 (2009).
Gineitis, A.A., Zalenskaya, I.A., Yau, P.M., Bradbury, E.M. & Zalensky, A.O. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J. Cell Biol. 151, 1591–1598 (2000).
Ku, M. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 (2008).
Chu, D.S. et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443, 101–105 (2006).
Dorus, S. et al. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445 (2006).
Ooi, S.L. & Henikoff, S. Germline histone dynamics and epigenetics. Curr. Opin. Cell Biol. 19, 257–265 (2007).
van der Heijden, G.W. et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 (2005).
Torres-Padilla, M.E., Bannister, A.J., Hurd, P.J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Minami, N., Suzuki, T. & Tsukamoto, S. Zygotic gene activation and maternal factors in mammals. J. Reprod. Dev. 53, 707–715 (2007).
Schuettengruber, B. & Cavalli, G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 (2009).
Bao, S. et al. Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep. 6, 748–754 (2005).
Kawahara, M. et al. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat. Biotechnol. 25, 1045–1050 (2007).
Ng, R.K. & Gurdon, J.B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10, 102–109 (2008).
Katz, D.J., Edwards, T.M., Reinke, V. & Kelly, W.G.A. C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308–320 (2009).
van der Heijden, G.W. et al. Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol. Reprod. Dev. 76, 101–108 (2009).
Ramos, L. et al. Incomplete nuclear transformation of human spermatozoa in oligo-astheno-teratospermia: characterization by indirect immunofluorescence of chromatin and thiol status. Hum. Reprod. 23, 259–270 (2008).
Chong, S. et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat. Genet. 39, 614–622 (2007).
Lee, K., Haugen, H.S., Clegg, C.H. & Braun, R.E. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc. Natl. Acad. Sci. USA 92, 12451–12455 (1995).
Dechat, T. et al. Detergent-salt resistance of LAP2α in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 17, 4887–4902 (1998).
O'Geen, H., Nicolet, C.M., Blahnik, K., Green, R. & Farnham, P.J. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41, 577–580 (2006).
Umlauf, D., Goto, Y. & Feil, R. Site-specific analysis of histone methylation and acetylation. Methods Mol. Biol. 287, 99–120 (2004).
Acknowledgements
We thank T. Jenuwein (Max-Planck Institute of Immunobiology, Freiburg, Germany) and R. Foisner (Max F. Perutz Laboratories, Medical University of Vienna, Vienna, Austria) for providing antisera. We are grateful to F. Zilbermann and the Friedrich Miescher Institute (FMI) animal facility for excellent technical assistance. We thank P. de Boer and R. Terranova for critical reading of the manuscript and all members of the Peters and Schübeler laboratories for discussions. U.B. and S.E. acknowledge the Boehringer Ingelheim Fonds for their PhD fellowships. M.H. was supported by the Swiss National Science Foundation and the Japan Society for the Promotion of Science. T.C.R. was supported by the Swiss Cancer Leagues. Research in the Peters laboratory is supported by the Novartis Research Foundation, the Swiss National Science Foundation, the European Network of Excellence “The Epigenome” and the EMBO YIP program.
Author information
Authors and Affiliations
Contributions
U.B., M.H., S.E. and A.H.F.M.P. conceived and designed the experiments; U.B., M.H. and S.E. performed the experiments; L.R. provided purified samples of human spermatozoa; U.B., M.H., S.E., E.J.O., M.B.S. and A.H.F.M.P. analyzed the data; C.B. contributed to deep-sequencing; T.C.R. and D.S. provided advice on data analyses and the manuscript; U.B., S.E. and A.H.F.M.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Methods (PDF 2592 kb)
Supplementary Table 1
List of genes with modification states in human spermatozoa (XLS 3667 kb)
Supplementary Table 2
List of significantly over- and under-represented GO terms for Fig. 1d, 3d, 3e (XLS 842 kb)
Supplementary Table 3
Real-time PCR primer sequences (XLS 38 kb)
Rights and permissions
About this article
Cite this article
Brykczynska, U., Hisano, M., Erkek, S. et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17, 679–687 (2010). https://doi.org/10.1038/nsmb.1821
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1821
This article is cited by
-
Mapping crossover events of mouse meiotic recombination by restriction fragment ligation-based Refresh-seq
Cell Discovery (2024)
-
Chromosome ends and the theory of marginotomy: implications for reproduction
Biogerontology (2024)
-
Sperm chromatin accessibility’s involvement in the intergenerational effects of stress hormone receptor activation
Translational Psychiatry (2023)
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Emerging evidence that the mammalian sperm epigenome serves as a template for embryo development
Nature Communications (2023)