Key Points
-
Small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs) serve as the effector molecules of RNA interference (RNAi). Important mechanistic advances have recently been made in understanding the diverse ways in which RNAi pathways regulate gene expression.
-
The potency and specificity of chemically synthesized siRNAs and expressed shRNAs are increased by designing these molecules so that they serve as substrates for the RNase III enzymes Drosha and Dicer. Expressed shRNAs can be designed to mimic the structures of endogenous microRNA (miRNA) transcripts.
-
Sequence-dependent off-target and immunostimulatory effects are concerns that must be addressed when designing siRNAs and shRNAs for therapeutic purposes. Saturation of RNAi pathway components should also be avoided by optimizing siRNA dosage and shRNA expression levels.
-
siRNAs are delivered systemically through lipid-based carriers and liganded nanoparticles. New methods have been developed that allow siRNAs to be targeted to specific cell-surface receptors.
-
Viral delivery vectors facilitate the stable expression of shRNAs in therapeutically relevant settings. Lentiviral, adenoviral and adeno-associated viral vectors are in development for RNAi gene-therapy strategies.
-
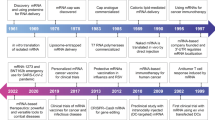
Clinical trials using RNAi-based therapies are currently under way for wet, age-related macular degeneration and respiratory syncytial virus infection. At present, RNAi-based therapies for other viral infections, cancer and neurodegenerative diseases are in preclinical development, and additional clinical trials will be initiated during 2007.
Abstract
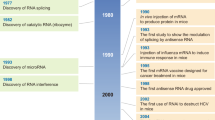
Since the first description of RNA interference (RNAi) in animals less than a decade ago, there has been rapid progress towards its use as a therapeutic modality against human diseases. Advances in our understanding of the mechanisms of RNAi and studies of RNAi in vivo indicate that RNAi-based therapies might soon provide a powerful new arsenal against pathogens and diseases for which treatment options are currently limited. Recent findings have highlighted both promise and challenges in using RNAi for therapeutic applications. Design and delivery strategies for RNAi effector molecules must be carefully considered to address safety concerns and to ensure effective, successful treatment of human diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Fire, A. et al. Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). This Nobel Prize-winning landmark paper provides the first description of the phenomenon of RNAi.
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001). This work provides the first description of RNAi in mammalian cells.
Hannon, G. J. & Rossi, J. J. Unlocking the potential of the human genome with RNA interference. Nature 431, 371–378 (2004).
Check, E. A crucial test. Nature Med. 11, 243–244 (2005).
McFarland, T. J., Zhang, Y., Appukuttan, B. & Stout, J. T. Gene therapy for proliferative ocular diseases. Expert Opin. Biol. Ther. 4, 1053–1058 (2004).
Bitko, V., Musiyenko, A., Shulyayeva, O. & Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nature Med. 11, 50–55 (2005).
Rossi, J. J. RNAi as a treatment for HIV-1 infection. Biotechniques 40, s25–s29 (2006).
Dykxhoorn, D. M. & Lieberman, J. Silencing viral infection. PLoS Med. 3, e242 (2006).
Raoul, C., Barker, S. D. & Aebischer, P. Viral-based modeling and correction of neurodegenerative diseases by RNA interference. Gene Ther. 13, 487–495 (2006).
Pai, S. I. et al. Prospects of RNA interference therapy for cancer. Gene Ther. 13, 464–477 (2006).
Amarzguioui, M., Rossi, J. J. & Kim, D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 579, 5974–5981 (2005).
de Veer, M. J., Sledz, C. A. & Williams, B. R. Detection of foreign RNA: implications for RNAi. Immunol. Cell Biol. 83, 224–228 (2005).
Grimm, D. et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006). This paper describes the in vivo toxicity of long-term, high-level shRNA expression in mice.
Behlke, M. A. Progress towards in vivo use of siRNAs. Mol. Ther. 13, 644–670 (2006).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004).
Matzke, M. A. & Birchler, J. A. RNAi-mediated pathways in the nucleus. Nature Rev. Genet. 6, 24–35 (2005).
Wassenegger, M. The role of the RNAi machinery in heterochromatin formation. Cell 122, 13–16 (2005).
Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 (2000).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 (2002).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Matranga, C., Tomari, Y., Shin, C., Bartel, D. P. & Zamore, P. D. Passenger-strand cleavage facilitates assembly of siRNA into AGO2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005).
Rand, T. A., Petersen, S., Du, F. & Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 (2005).
Parker, J. S., Roe, S. M. & Barford, D. Structural insights into mRNA recognition from a PIWI domain–siRNA guide complex. Nature 434, 663–666 (2005).
Ma, J. B., Ye, K. & Patel, D. J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322 (2004).
Elbashir, S. M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Orban, T. I. & Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11, 459–469 (2005).
Hutvagner, G. & Zamore, P. D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 (2002).
Bartel, D. P. & Chen, C. Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Rev. Genet. 5, 396–400 (2004).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Liu, J., Valencia-Sanchez, M. A., Hannon, G. J. & Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biol. 7, 719–723 (2005).
Yekta, S., Shih, I. H. & Bartel, D. P. MicroRNA-directed cleavage of H oxb8 mRNA. Science 304, 594–596 (2004).
Lee, Y. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 (2004).
Borchert, G. M., Lanier, W. & Davidson, B. L. RNA polymerase III transcribes human microRNAs. Nature Struct. Mol. Biol. 13, 1097–1101 (2006).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003).
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95–98 (2004).
Chendrimada, T. P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005).
Lee, Y. et al. The role of PACT in the RNA silencing pathway. EMBO J. 25, 522–532 (2006).
Preall, J. B. & Sontheimer, E. J. RNAi: RISC gets loaded. Cell 123, 543–245 (2005).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Silva, J. M. et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genet. 37, 1281–1288 (2005).
Morris, K. V., Chan, S. W., Jacobsen, S. E. & Looney, D. J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 (2004).
Kim, D. H., Villeneuve, L. M., Morris, K. V. & Rossi, J. J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nature Struct. Mol. Biol. 13, 793–797 (2006).
Janowski, B. A. et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nature Struct. Mol. Biol. 13, 787–792 (2006).
Ting, A. H., Schuebel, K. E., Herman, J. G. & Baylin, S. B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nature Genet. 37, 906–910 (2005).
Weinberg, M. S. et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12, 256–262 (2006).
Vastenhouw, N. L. et al. Gene expression: long-term gene silencing by RNAi. Nature 442, 882 (2006).
Kim, D. H. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature Biotechnol. 23, 222–226 (2005).
Siolas, D. et al. Synthetic shRNAs as potent RNAi triggers. Nature Biotechnol. 23, 227–231 (2005).
Marques, J. T. et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nature Biotechnol. 24, 559–565 (2006).
Brummelkamp, T. R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958 (2002). References 54 and 55 describe stable expression systems for RNAi.
Zeng, Y., Wagner, E. J. & Cullen, B. R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9, 1327–1333 (2002).
Sarnow, P., Jopling, C. L., Norman, K. L., Schutz, S. & Wehner, K. A. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nature Rev. Microbiol. 4, 651–659 (2006).
Jackson, A. L. et al. Widespread siRNA 'off-target' transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 (2006).
Birmingham, A. et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods 3, 199–204 (2006).
Hornung, V. et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Med. 11, 263–270 (2005). This work describes the detection of immunostimulatory motifs in siRNA sequences through TLRs.
Manche, L., Green, S. R., Schmedt, C. & Mathews, M. B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12, 5238–5248 (1992).
Seth, R. B., Sun, L. & Chen, Z. J. Antiviral innate immunity pathways. Cell Res. 16, 141–147 (2006).
Judge, A. D. et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnol. 23, 457–462 (2005).
Kim, D. H. et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nature Biotechnol. 22, 321–325 (2004).
Robbins, M. A. et al. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nature Biotechnol. 24, 566–571 (2006).
An, D. S. et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 14, 494–504 (2006).
Czauderna, F. et al. Structural variations and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31, 2705–2716 (2003).
Morrissey, D. V. et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41, 1349–1356 (2005).
Chiu, Y. L. & Rana, T. M. siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003).
Morrissey, D. V. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotechnol. 23, 1002–1007 (2005).
Soutschek, J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004). This work describes a straightforward and effective method for intravenous systemic delivery of RNAi.
Zimmermann, T. S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006). This paper describes the potency and longevity of systemic delivery of RNAi in non-human primates.
Song, E. et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnol. 23, 709–717 (2005).
McNamara, J. O. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nature Biotechnol. 24, 1005–1015 (2006).
Chu, T. C., Twu, K. Y., Ellington, A. D. & Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 34, e73 (2006).
Hu-Lieskovan, S., Heidel, J. D., Bartlett, D. W., Davis, M. E. & Triche, T. J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 65, 8984–8992 (2005).
Morris, K. V. & Rossi, J. J. Lentiviral-mediated delivery of siRNAs for antiviral therapy. Gene Ther. 13, 553–558 (2006).
Grimm, D. & Kay, M. A. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 13, 563–575 (2006).
Lee, N. S. et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nature Biotechnol. 20, 500–505 (2002).
Novina, C. D. et al. siRNA-directed inhibition of HIV-1 infection. Nature Med. 8, 681–686 (2002).
Jacque, J., Triques, K. & Stevenson, M. Modulation of HIV-1 replication by RNA interference. Nature 418, 435–438 (2002).
Coburn, G. A. & Cullen, B. R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76, 9225–9231 (2002).
Martinez, J. & Tuschl, T. RISC is a 5' phosphomonoester-producing RNA endonuclease. Genes Dev. 18, 975–980 (2004).
Huang, Y. et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nature Med. 2, 1240–1243 (1996).
Li, M. J. et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 12, 900–909 (2005).
Zhang, W. et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nature Med. 11, 56–62 (2005).
Palliser, D. et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439, 89–94 (2006). This paper describes the efficacy of topical delivery of RNAi as a microbicide.
Xia, H. et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nature Med. 10, 816–820 (2004).
Raoul, C. et al. Lentiviral-mediated silencing of Sod1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Med. 11, 423–428 (2005).
Ralph, G. S. et al. Silencing mutant Sod1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nature Med. 11, 429–433 (2005).
Pai, S. I. et al. Prospects of RNA interference therapy for cancer. Gene Ther. 13, 464–477 (2006).
Landen, C. N. et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 65, 6910–6918 (2005).
Dykxhoorn, D. M. & Lieberman, J. Knocking down disease with siRNAs. Cell 126, 231–235 (2006).
Acknowledgements
We thank members of our laboratory for discussions and support. We apologize to any colleagues whose work could not be cited due to space limitations. This work was supported by grants from the US National Institutes of Health to J.J.R.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Daniel H. Kim & John J. Rossi. Strategies for silencing human disease using RNA interference. Nature Reviews Genetics 8, 173–184 (2007); doi:10.1038/nrg2006
John J. Rossi is a co-founder and equity holder in Calando Pharmaceuticals, Inc. (Pasadena, California, USA) — an RNAi company.
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
Ambion RNAi Interference Resource
Integrated DNA Technologies RNAi design tools
Glossary
- Short hairpin RNAs
-
(shRNAs). A class of small RNAs with a stem of 19–29 base pairs and a loop of 4–10 nucleotides that are processed by Dicer into small interfering RNAs. shRNAs are expressed from vectors to induce RNAi.
- Interferons
-
A class of glycoproteins that are upregulated in response to exogenous ssRNA or dsRNA as a cellular defence mechanism against RNA viral infection.
- Processing bodies
-
Cytoplasmic bodies that contain enzymes involved in mRNA turnover, such as the decapping enzymes DCP1 and DCP2, and sequester mRNAs from the translational machinery.
- Polycistronic
-
A single RNA molecule that can generate several products. For miRNAs, a single polycistronic transcript contains multiple stem-loop structures encoding separate miRNAs.
- Plasmacytoid dendritic cells
-
Cells of the immune system that recognize foreign pathogens through Toll-like receptors and other pattern-recognition receptors.
- Endosome
-
A vesicle formed during the incorporation of extracellular material by endocytosis. Toll-like receptors are found in endosomal compartments.
- Nanoparticle
-
Nanometre-scale particles that are formulated from polymers or phospholipids and are used as delivery vehicles for therapeutic applications.
- Aptamer
-
RNA or DNA oligonucleotides selected from random pools of sequences that bind to specific receptors on the basis of their secondary structure.
- Episome
-
A double-stranded, circular DNA molecule that replicates in the nucleus without integrating into the host genome.
- Pseudotyping
-
Changing the ability of a viral vector to bind cell-surface receptors by altering its envelope proteins.
- Hammerhead ribozyme
-
Small, self-cleaving catalytic RNAs with distinct secondary structures and highly conserved core residues that mediate cleavage.
Rights and permissions
About this article
Cite this article
Kim, D., Rossi, J. Strategies for silencing human disease using RNA interference. Nat Rev Genet 8, 173–184 (2007). https://doi.org/10.1038/nrg2006
Issue Date:
DOI: https://doi.org/10.1038/nrg2006
This article is cited by
-
Biotechnological strategies to decipher the functions of abiotic stress-associated genes in soybean
Plant Biotechnology Reports (2024)
-
Advances in siRNA delivery approaches in cancer therapy: challenges and opportunities
Molecular Biology Reports (2023)
-
Current Insights of Nanocarrier-Mediated Gene Therapeutics to Treat Potential Impairment of Amyloid Beta Protein and Tau Protein in Alzheimer’s Disease
Molecular Neurobiology (2023)
-
Emerging trends in the nanomedicine applications of functionalized magnetic nanoparticles as novel therapies for acute and chronic diseases
Journal of Nanobiotechnology (2022)
-
The inhibitory role of si-UBB delivered by degradable dendrimers-based lipid nanoparticles in ovarian cancer
Cancer Nanotechnology (2022)