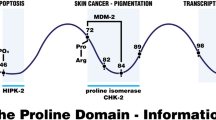
Abstract
The gene TP53, encoding p53, has a common sequence polymorphism that results in either proline or arginine at amino-acid position 72. This polymorphism occurs in the proline-rich domain of p53, which is necessary for the protein to fully induce apoptosis. We found that in cell lines containing inducible versions of alleles encoding the Pro72 and Arg72 variants, and in cells with endogenous p53, the Arg72 variant induces apoptosis markedly better than does the Pro72 variant. Our data indicate that at least one source of this enhanced apoptotic potential is the greater ability of the Arg72 variant to localize to the mitochondria; this localization is accompanied by release of cytochrome c into the cytosol. These data indicate that the two polymorphic variants of p53 are functionally distinct, and these differences may influence cancer risk or treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
el-Deiry, W.S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993).
Deng, C., Zhang, P., Harper, J.W., Elledge, S.J. & Leder, P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82, 675–684 (1995).
Caelles, C., Helmberg, A. & Karin, M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370, 220–223 (1994).
Wagner, A.J., Kokontis, J.M. & Hay, N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 8, 2817–2830 (1994).
Koumenis, C. et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21, 1297–1310 (2001).
Haupt, Y., Rowan, S., Shaulian, E., Vousden, K.H. & Oren, M. Induction of apoptosis in HeLa cells by transactivation-deficient p53. Genes Dev. 9, 2170–2183 (1995).
Walker, K.K. & Levine, A.J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. USA 93, 15335–15340 (1996).
Sakamuro, D., Sabbatini, P., White, E. & Prendergast, G.C. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15, 887–898 (1997).
Buchman, V.L., Chumakov, N.N., Ninkina, N.N., Samarina, O.P. & Georgiev, G.P. A variation in the structure of the protein-coding region of the human p53 gene. Gene 70, 245–252 (1988).
Harris, N. et al. Molecular basis for heterogeneity of the human p53 protein. Mol Cell. Biol. 6, 4650–4656 (1986).
Matlashewski, G.J. et al. Primary structure polymorphism at amino acid residue 72 of human p53. Mol. Cell. Biol. 7, 961–963 (1987).
Sjalander, A., Birgander, R., Kivela, A. & Beckman, G. p53 polymorphisms and haplotypes in different ethnic groups. Hum. Hered. 45, 144–149 (1995).
Beckman, G. et al. Is p53 polymorphism maintained by natural selection? Hum. Hered. 44, 266–270 (1994).
Thomas, M. et al. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol. Cell. Biol. 19, 1092–1100 (1999).
Storey, A. et al. Role of a p53 polymorphism in the development of human papilloma-virus-associated cancer. Nature 393, 229–234 (1998).
Marin, M.C. et al. A common polymorphism acts as an intragenic modifier of mutant p53 behavior. Nat. Genet. 25, 47–54 (2000).
Pochampally, R. et al. Temperature-sensitive mutants of p53 homologs. Biochem. Biophys. Res. Commun. 279, 1001–1010 (2000).
Wistuba, I.I., Gazdar, A.F. & Minna, J.D. Molecular genetics of small cell lung carcinoma. Seminars Oncol. 28, 3–13 (2001).
Landers, J.E., Cassel, S.L. & George, D.L. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 57, 3562–3568 (1997).
Attardi, L.D. et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14, 704–718 (2000).
Marchenko, N.D., Zaika, A. & Moll, U.M. Death signal–induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212 (2000).
Sansome, C., Zaika, A., Marchenko, N.D. & Moll, U.M. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 488, 110–115 (2001).
Pfanner, N. & Geissler, A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349 (2001).
Zilfou, J.T., Hoffman, W.H., Sank, M., George, D.L. & Murphy, M. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21, 3974–3985 (2001).
Boyd, S.D., Tsai, K.Y. & Jacks, T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell. Biol. 2, 563–568 (2000).
Geyer, R.K., Yu, Z.K. & Maki, C.G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell. Biol. 2, 569–573 (2000).
Chen, L., Marechal, V., Moreau, J., Levine, A.J. & Chen, J. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J. Biol. Chem. 272, 22966–22973 (1997).
Berger, M., Sionov, R.V., Levine, A.J. & Haupt, Y. A role for the polyproline domain of p53 in its regulation by MDM2. J. Biol. Chem. 276, 3785–3790 (2001).
Hicke, L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 (2001).
Chen, G. et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J. Biol. Chem. 274, 7–10 (1999).
Daugas, E. et al. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 14, 729–739 (2000).
Samali, A., Cai, J., Zhivotovsky, B., Jones, D.P. & Orrenius, S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of Jurkat cells. EMBO J. 18, 2040–2048 (1999).
Xanthoudakis, S. et al. Hsp60 accelerates the maturation of pro-caspase-3 by upstream activator proteases during apoptosis. EMBO J. 18, 2049–2056 (1999).
Weston, A. et al. Determination of the allelic frequencies of an L-myc and a p53 polymorphism in human lung cancer. Carcinogenesis 15, 583–587 (1994).
Birgander, R. et al. p53 polymorphisms and haplotypes in lung cancer. Carcinogenesis 16, 2233–2236 (1995).
Rosenthal, A. et al. p53 codon 72 polymorphism and risk of cervical cancer in UK. Lancet 352, 871–872 (1998).
Acknowledgements
The authors would like to thank J. Skipworth for technical assistance, A. Ganguly and C. Spittle for genotyping the melanoma and fibroblast cell lines and J. Boyd for confocal expertise. This work was supported by US Public Health Service National Cancer Institute grants to D.L.G. and M.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dumont, P., Leu, JJ., Della Pietra, A. et al. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33, 357–365 (2003). https://doi.org/10.1038/ng1093
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1093
This article is cited by
-
Association of TP53 rs1042522 G > C, MDM2 rs2279744 T > G, and miR-34b/c rs4938723 T > C polymorphisms with aneuploidy pregnancy susceptibility
BMC Pregnancy and Childbirth (2023)
-
Genetic variability in cisplatin metabolic pathways and outcome of locally advanced head and neck squamous cell carcinoma patients
Scientific Reports (2023)
-
Targeting PSAT1 to mitigate metastasis in tumors with p53-72Pro variant
Signal Transduction and Targeted Therapy (2023)
-
TP53 mutations in functional corticotroph tumors are linked to invasion and worse clinical outcome
Acta Neuropathologica Communications (2022)
-
Association of TP53 gene polymorphisms with the risk of acute lymphoblastic leukemia in Moroccan children
Molecular Biology Reports (2022)