Abstract
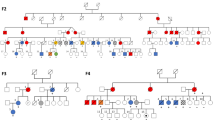
We report the discovery of GATA2 as a new myelodysplastic syndrome (MDS)-acute myeloid leukemia (AML) predisposition gene. We found the same, previously unidentified heterozygous c.1061C>T (p.Thr354Met) missense mutation in the GATA2 transcription factor gene segregating with the multigenerational transmission of MDS-AML in three families and a GATA2 c.1063_1065delACA (p.Thr355del) mutation at an adjacent codon in a fourth MDS family. The resulting alterations reside within the second zinc finger of GATA2, which mediates DNA-binding and protein-protein interactions. We show differential effects of the mutations on the transactivation of target genes, cellular differentiation, apoptosis and global gene expression. Identification of such predisposing genes to familial forms of MDS and AML is critical for more effective diagnosis and prognosis, counseling, selection of related bone marrow transplant donors and development of therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Vardiman, J.W. et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114, 937–951 (2009).
Barzi, A. & Sekeres, M.A. Myelodysplastic syndromes: a practical approach to diagnosis and treatment. Cleve. Clin. J. Med. 77, 37–44 (2010).
Owen, C., Barnett, M. & Fitzgibbon, J. Familial myelodysplasia and acute myeloid leukaemia–a review. Br. J. Haematol. 140, 123–132 (2008).
Zhang, P. et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA 96, 8705–8710 (1999).
Crossley, M., Merika, M. & Orkin, S.H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15, 2448–2456 (1995).
Zhang, S.J. et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA 105, 2076–2081 (2008).
Yan, X.J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315 (2011).
Usary, J. et al. Mutation of GATA3 in human breast tumors. Oncogene 23, 7669–7678 (2004).
Owen, C.J. et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 112, 4639–4645 (2008).
Carmichael, C.L. et al. Poor prognosis in familial acute myeloid leukaemia with combined biallelic CEBPA mutations and downstream events affecting the ATM, FLT3 and CDX2 genes. Br. J. Haematol. 150, 382–385 (2010).
Robinson, C.W., Norman, J.E., Cleland, L.G. & Ford, J.H. Dermal necrosis and chromosome Iq abnormality in a man with a familial myeloproliferative disorder. Aust. N. Z. J. Med. 13, 141–145 (1983).
Horwitz, M., Sabath, D.E., Smithson, W.A. & Radich, J. A family inheriting different subtypes of acute myelogenous leukemia. Am. J. Hematol. 52, 295–304 (1996).
Minelli, A. et al. Familial partial monosomy 7 and myelodysplasia: different parental origin of the monosomy 7 suggests action of a mutator gene. Cancer Genet. Cytogenet. 124, 147–151 (2001).
Michaud, J. et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics 9, 363 (2008).
Veitia, R.A. Dominant negative factors in health and disease. J. Pathol. 218, 409–418 (2009).
Ernst, T. et al. Transcription factor mutations in myelodysplastic/myeloproliferative neoplasms. Haematologica 95, 1473–1480 (2010).
Matheny, C.J. et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 26, 1163–1175 (2007).
Kitajima, K. et al. Redirecting differentiation of hematopoietic progenitors by a transcription factor, GATA-2. Blood 107, 1857–1863 (2006).
Tsai, F.Y. et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994).
Zhang, S.J. & Shi, J.Y. GATA-2 L359V mutation is solely associated with CML progression but not other hematological malignancies. Blood 112, Abstract 1507 (2008).
Zhang, S.J., Shi, J.Y. & Li, J.Y. GATA-2 L359V mutation is exclusively associated with CML progression but not other hematological malignancies and GATA-2 P250A is a novel single nucleotide polymorphism. Leuk. Res. 33, 1141–1143 (2009).
Fadilah, S.A., Cheong, S.K., Roslan, H., Rozie-Hanisa, M. & Yen, G.K. GATA-1 and GATA-2 gene expression is related to the severity of dysplasia in myelodysplastic syndrome. Leukemia 16, 1563–1565 (2002).
Bullinger, L. et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 24, 438–449 (2010).
Wieser, R. et al. Transcription factor GATA-2 gene is located near 3q21 breakpoints in myeloid leukemia. Biochem. Biophys. Res. Commun. 273, 239–245 (2000).
Bullinger, L. et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 350, 1605–1616 (2004).
Lahortiga, I. et al. Molecular heterogeneity in AML/MDS patients with 3q21q26 rearrangements. Genes Chromosom. Cancer 40, 179–189 (2004).
Slape, C. et al. Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res. 67, 5148–5155 (2007).
Hsu, A.P. et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood published online, doi:10.1182/blood-2011-05-356352 (13 June 2011).
Dickinson, R.E. et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood published online, doi:10.1182/blood-2011-06-360313 (15 July 2011).
Mansour, S. et al. Emberger syndrome-primary lymphedema with myelodysplasia: report of seven new cases. Am. J. Med. Genet. A 152A, 2287–2296 (2010).
Ostergaard, P. et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat. Genet. advance online publication, doi:10.1038/ng.923 (4 September 2011).
Tsuzuki, S., Towatari, M., Saito, H. & Enver, T. Potentiation of GATA-2 activity through interactions with the promyelocytic leukemia protein (PML) and the t(15;17)-generated PML-retinoic acid receptor alpha oncoprotein. Mol. Cell. Biol. 20, 6276–6286 (2000).
Diessenbacher, P. et al. NF-κB inhibition reveals differential mechanisms of TNF versus TRAIL-induced apoptosis upstream or at the level of caspase-8 activation independent of cIAP2. J. Invest. Dermatol. 128, 1134–1147 (2008).
Callus, B.A. et al. Triggering of apoptosis by Puma is determined by the threshold set by prosurvival Bcl-2 family proteins. J. Mol. Biol. 384, 313–323 (2008).
Koldej, R., Cmielewski, P., Stocker, A., Parsons, D.W. & Anson, D.S. Optimisation of a multipartite human immunodeficiency virus based vector system; control of virus infectivity and large-scale production. J. Gene Med. 7, 1390–1399 (2005).
Kumar, R. et al. CBFA2T3-ZNF652 corepressor complex regulates transcription of the E-box gene HEB. J. Biol. Chem. 283, 19026–19038 (2008).
Acknowledgements
The authors would like to thank the families for their participation in our studies. Thanks also to J. Melo for provision of CML cell lines and R. Sharma for help with EMSA. Large-scale Sanger sequencing was performed by the Australian Genome Research Facility, which was established through the Commonwealth-funded Major National Research Facilities program. This work was supported by grants from the National Health and Medical Research Council of Australia (program grants 257501 (H.S.S.) and 219176 (H.S.S.), project grant 1002317 (C.N.H., R.J.D. and H.S.S.), fellowships 171601 (H.S.S.) and 461204 (H.S.S.), and a Dora Lush Postgraduate Award (C.L.C.)), Leukaemia Foundation of Australia (grant in aid to H.S.S. and a postdoctoral fellowship to C.L.C.), the Cancer Council of South Australia (H.S.S.) and MedVet Pty Ltd (H.S.S. and R.J.D.), Adelaide Scholarships International (scholarship to C.-E.C.) and US National Institutes of Health grant R01DK058161 (M.S.H.).
Author information
Authors and Affiliations
Contributions
C.N.H., R.J.D., M.S.H. and H.S.S. managed the project. C.N.H., C.L.C., E.J.W., C.-E.C., P.J.B., X.-C.L., M.B., M.L., A.C., Y.K.L., C.M.B., K.L.F. and A.E.T. performed the experiments. C.H.K. and R.J.D. performed structural modeling, and C.N.H., C.H.K. and L.G. performed data analysis. L.B.T., M.A., J.S., P.G.B., G.K.S., R.J.D., M.S.H. and H.S.S. collected families with MDS and/or AML and provided clinical data and samples. R.E. and P.G.E. participated in experimental design and provided critical reagents. A.L.B., I.D.L., S.M. and L.B.T. provided sporadic AML samples and correlative clinical data. C.N.H., R.J.D., M.S.H. and H.S.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–13 and Supplementary Note. (PDF 1327 kb)
Rights and permissions
About this article
Cite this article
Hahn, C., Chong, CE., Carmichael, C. et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 43, 1012–1017 (2011). https://doi.org/10.1038/ng.913
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.913
This article is cited by
-
The International Consensus Classification (ICC) of hematologic neoplasms with germline predisposition, pediatric myelodysplastic syndrome, and juvenile myelomonocytic leukemia
Virchows Archiv (2023)
-
Clonal evolution in leukemia: preleukemia, evolutionary models, and clinical implications
Genome Instability & Disease (2023)
-
Heterozygous variants in GATA2 contribute to DCML deficiency in mice by disrupting tandem protein binding
Communications Biology (2022)
-
Integrated stem cell signature and cytomolecular risk determination in pediatric acute myeloid leukemia
Nature Communications (2022)
-
A Nationwide Study of GATA2 Deficiency in Norway—the Majority of Patients Have Undergone Allo-HSCT
Journal of Clinical Immunology (2022)