Abstract
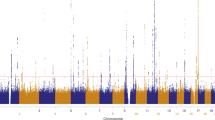
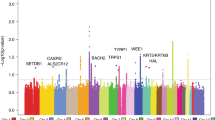
A high melanocytic nevi count is the strongest known risk factor for cutaneous melanoma. We conducted a genome-wide association study for nevus count using 297,108 SNPs in 1,524 twins, with validation in an independent cohort of 4,107 individuals. We identified strongly associated variants in MTAP, a gene adjacent to the familial melanoma susceptibility locus CDKN2A on 9p21 (rs4636294, combined P = 3.4 × 10−15), as well as in PLA2G6 on 22q13.1 (rs2284063, combined P = 3.4 × 10−8). In addition, variants in these two loci showed association with melanoma risk in 3,131 melanoma cases from two independent studies, including rs10757257 at 9p21, combined P = 3.4 × 10−8, OR = 1.23 (95% CI = 1.15–1.30) and rs132985 at 22q13.1, combined P = 2.6 × 10−7, OR = 1.23 (95% CI = 1.15–1.30). This provides the first report of common variants associated to nevus number and demonstrates association of these variants with melanoma susceptibility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Karim-Kos, H.E. et al. Recent trends of cancer in Europe: a combined approach of incidence, survival and mortality for 17 cancer sites since the 1990s. Eur. J. Cancer 44, 1345–1389 (2008).
Goldstein, A.M. et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J. Med. Genet. 44, 99–106 (2007).
Palmieri, G. et al. Definition of the role of chromosome 9p21 in sporadic melanoma through genetic analysis of primary tumours and their metastases. The Melanoma Cooperative Group. Br. J. Cancer 83, 1707–1714 (2000).
Fountain, J.W., Bale, S.J., Housman, D.E. & Dracopoli, N.C. Genetics of melanoma. Cancer Surv. 9, 645–671 (1990).
Gandini, S. et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer 41, 28–44 (2005).
Falchi, M., Spector, T.D., Perks, U., Kato, B.S. & Bataille, V. Genome-wide search for nevus density shows linkage to two melanoma loci on chromosome 9 and identifies a new QTL on 5q31 in an adult twin cohort. Hum. Mol. Genet. 15, 2975–2979 (2006).
Zhu, G. et al. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur. J. Hum. Genet. 15, 94–102 (2007).
Ma, Z., Wang, X., Nowatzke, W., Ramanadham, S. & Turk, J. Human pancreatic islets express mRNA species encoding two distinct catalytically active isoforms of group VI phospholipase A2 (iPLA2) that arise from an exon-skipping mechanism of alternative splicing of the transcript from the iPLA2 gene on chromosome 22q13.1. J. Biol. Chem. 274, 9607–9616 (1999).
Garcia-Castellano, J.M. et al. Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin. Cancer Res. 8, 782–787 (2002).
Hori, Y. et al. The methylthioadenosine phosphorylase gene is frequently co-deleted with the p16INK4a gene in acute type adult T-cell leukemia. Int. J. Cancer 75, 51–56 (1998).
Christopher, S.A., Diegelman, P., Porter, C.W. & Kruger, W.D. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 62, 6639–6644 (2002).
Behrmann, I. et al. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am. J. Pathol. 163, 683–690 (2003).
Pasmant, E. et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969 (2007).
Easton, D.F. & Eeles, R.A. Genome-wide association studies in cancer. Hum. Mol. Genet. 17, R109–R115 (2008).
Hosgood, H.D. III et al. Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis 29, 1938–1943 (2008).
Hooks, S.B. & Cummings, B.S. Role of Ca2+-independent phospholipase A2 in cell growth and signaling. Biochem. Pharmacol. 76, 1059–1067 (2008).
Atsumi, G. et al. Fas-induced arachidonic acid release is mediated by Ca2+-independent phospholipase A2 but not cytosolic phospholipase A2, which undergoes proteolytic inactivation. J. Biol. Chem. 273, 13870–13877 (1998).
Bao, S. et al. Effects of stable suppression of Group VIA phospholipase A2 expression on phospholipid content and composition, insulin secretion, and proliferation of INS-1 insulinoma cells. J. Biol. Chem. 281, 187–198 (2006).
Song, Y. et al. Inhibition of calcium-independent phospholipase A2 suppresses proliferation and tumorigenicity of ovarian carcinoma cells. Biochem. J. 406, 427–436 (2007).
Sun, B., Zhang, X., Talathi, S. & Cummings, B.S. Inhibition of Ca2+-independent phospholipase A2 decreases prostate cancer cell growth by p53-dependent and independent mechanisms. J. Pharmacol. Exp. Ther. 326, 59–68 (2008).
Bishop, D.T. et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet advance online publication, doi:10.1038/ng.411 (5 July 2009).
Bataille, V., Snieder, H., MacGregor, A.J., Sasieni, P. & Spector, D.T. Genetics of risk factors for melanoma: an adult twin study of nevi and freckles. J. Natl. Cancer Inst. 92, 457–463 (2000).
Sturm, R.A. et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 82, 424–431 (2008).
Baxter, A.J. et al. The Queensland Study of Melanoma: environmental and genetic associations (Q-MEGA); study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res. Hum. Genet. 11, 183–196 (2008).
Richards, J.B. et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet 371, 1505–1512 (2008).
Zhao, Z.Z. et al. Genetic variation in tumour necrosis factor and lymphotoxin is not associated with endometriosis in an Australian sample. Hum. Reprod. 22, 2389–2397 (2007).
Chen, W.M. & Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81, 913–926 (2007).
Devlin, B., Roeder, K. & Wasserman, L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 60, 155–166 (2001).
Lange, K., Weeks, D. & Boehnke, M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet. Epidemiol. 5, 471–472 (1988).
Saxena, R. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Prokopenko, I. et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 41, 77–81 (2009).
Acknowledgements
The KCL authors acknowledge financial support from the Wellcome Trust, the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London, EC Framework 7 Health-2007-A ENGAGE project and the Chronic Disease Research Foundation (CDRF). T.D.S. is an NIHR senior investigator. We would also like to thank all the nurses and research assistants who collected the skin data, including U. Perks, G. Clement, D. Glass and E. Qweitin as well as the volunteer twins who gave their time.
The Leeds case-control study and genotyping was supported by Cancer Research UK Programme Award (C588/A4994), NIH (R01 CA83115) and EU FP6 to GenoMEL (LSHC-CT-2006-018702).
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data are available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. The Leeds second control series were recruited through an award from the Department of Health in conjunction with I. dos Santos Silva and A. Swerdlow. We would like to thank J. Taylor for statistical analyses relating to the Leeds dataset. T.P. holds a Canada Research Chair and is supported by Genome Canada/Quebec and the CIHR.
The Australian Studies were supported by the National Institutes of Health/National Cancer Institute (CA88363), the National Health and Medical Research Council of Australia (NHMRC) and the Cancer Council Queensland. D.L.D., G.W.M. and N.K.H. are supported by the NHMRC Fellowships scheme. We thank D. Whiteman, A. Green, J. Aitken, A. Henders, M. Campbell, M. Stark, A. Baxter, M. de Nooyer, I. Gardner, D. Statham, B. Haddon, J. Palmer, L. Bardsley, D. Smyth and H. Beeby for their input into project management, sample processing, database and questionnaire development. We are grateful to all the participants of the BTNS and Q-MEGA as well as the research interviewers and examiners for these studies.
Author information
Authors and Affiliations
Contributions
M.F., T.D.S., N.G.M., D.T.B. and N.K.H. designed the study. M.F., D.L.D., J.H.B. and A.C. analysed the data. V.B., N.G.M. and J.A.N.B. contributed to data collection and phenotype definitions. T.D.S., P.D., D.E.E., N.G.M., N.K.H. and J.A.N.B. obtained funding. Z.Z.Z., P.D., N.S. and G.W.M. contributed to genotyping. M.F., V.B. and T.D.S. wrote the first draft of the paper. All authors made important contributions to the final version of the paper.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Tables 1–5 (PDF 352 kb)
Rights and permissions
About this article
Cite this article
Falchi, M., Bataille, V., Hayward, N. et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet 41, 915–919 (2009). https://doi.org/10.1038/ng.410
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.410
This article is cited by
-
Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma
Nature Communications (2020)
-
Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility
Nature Genetics (2020)
-
Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways
Nature Communications (2018)
-
Melanocytic nevi and melanoma: unraveling a complex relationship
Oncogene (2017)
-
New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk
Nature Communications (2016)