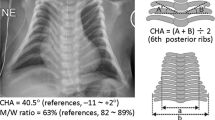
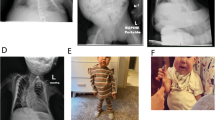
Abstract
Human chromosome 14q32.2 carries a cluster of imprinted genes including paternally expressed genes (PEGs) such as DLK1 and RTL1 and maternally expressed genes (MEGs) such as MEG3 (also known as GTL2), RTL1as (RTL1 antisense) and MEG8 (refs. 1,2), together with the intergenic differentially methylated region (IG-DMR) and the MEG3-DMR3,4,5. Consistent with this, paternal and maternal uniparental disomy for chromosome 14 (upd(14)pat and upd(14)mat) cause distinct phenotypes6,7. We studied eight individuals (cases 1–8) with a upd(14)pat-like phenotype and three individuals (cases 9–11) with a upd(14)mat-like phenotype in the absence of upd(14) and identified various deletions and epimutations affecting the imprinted region. The results, together with recent mouse data4,8,9,10, imply that the IG-DMR has an important cis-acting regulatory function on the maternally inherited chromosome and that excessive RTL1 expression and decreased DLK1 and RTL1 expression are relevant to upd(14)pat-like and upd(14)mat-like phenotypes, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cavaille, J., Seitz, H., Paulsen, M., Ferguson-Smith, A.C. & Bachellerie, J.P. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 11, 1527–1538 (2002).
Charlier, C. et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11, 850–862 (2001).
Paulsen, M. et al. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2–H19 region. Genome Res. 11, 2085–2094 (2001).
Lin, S.P. et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97–102 (2003).
Murphy, S.K. et al. Epigenetic detection of human chromosome 14 uniparental disomy. Hum. Mutat. 22, 92–97 (2003).
Kagami, M. et al. Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am. J. Med. Genet. A. 138, 127–132 (2005).
Kotzot, D. Maternal uniparental disomy 14 dissection of the phenotype with respect to rare autosomal recessively inherited traits, trisomy mosaicism, and genomic imprinting. Ann. Genet. 47, 251–260 (2004).
Sekita, Y. et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. advance online publication, 10.1038/ng.2007.51 (6 January 2008).
Seitz, H. et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262 (2003).
Davis, E. et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749 (2005).
Takahashi, I., Takahashi, T., Utsunomiya, M., Takada, G. & Koizumi, A. Long-acting gonadotropin-releasing hormone analogue treatment for central precocious puberty in maternal uniparental disomy chromosome 14. Tohoku J. Exp. Med. 207, 333–338 (2005).
Rosa, A.L. et al. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res. 13, 809–818 (2005).
Kosaki, K. et al. Diagnosis of maternal uniparental disomy of chromosome 7 with a methylation specific PCR assay. J. Med. Genet. 37, E19 (2000).
Gicquel, C. et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 37, 1003–1007 (2005).
Kubota, T. et al. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 16, 16–17 (1997).
Kaneko-Ishino, T., Kohda, T. & Ishino, F. The regulation and biological significance of genomic imprinting in mammals. J. Biochem. 133, 699–711 (2003).
Coan, P.M., Burton, G.J. & Ferguson-Smith, A.C. Imprinted genes in the placenta—a review. Placenta 26 (Suppl. A), S10–S20 (2005).
Kaneko-Ishino, T., Kohda, T., Ono, R. & Ishino, F. Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet. Genome Res. 113, 24–30 (2006).
Takada, S. et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2–H19. Hum. Mol. Genet. 11, 77–86 (2002).
Georgiades, P., Watkins, M., Surani, M.A. & Ferguson-Smith, A.C. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127, 4719–4728 (2000).
Lin, S.P. et al. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development 134, 417–426 (2007).
Moon, Y.S. et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 22, 5585–5592 (2002).
Temple, I.K. et al. Isolated imprinting mutation of the DLK1/GTL2 locus associated with a clinical presentation of maternal uniparental disomy of chromosome 14. J. Med. Genet. 44, 637–640 (2007) published online 29 June 2007.
Hernandez, A., Martinez, M.E., Fiering, S., Galton, V.A. & St Germain, D.L. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J. Clin. Invest. 116, 476–484 (2006).
Tsai, C.E. et al. Genomic imprinting contributes to thyroid hormone metabolism in the mouse embryo. Curr. Biol. 12, 1221–1226 (2002).
Sekita, Y. et al. Aberrant regulation of imprinted gene expression in Gtl2lacZ mice. Cytogenet. Genome Res. 113, 223–229 (2006).
Lewis, A. et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36, 1291–1295 (2004).
Umlauf, D. et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36, 1296–1300 (2004).
Acknowledgements
We would like to thank all the affected individuals and their family members who participated in this study. This work was supported by Grants for Child Health and Development (17C-2) and for Research on Children and Families (H18-005) from the Ministry of Health, Labor, and Welfare, and by Grants-in-Aid for Scientific Research (priority areas: 16086215 and 1508023; category B: 19390290) from the Ministry of Education, Culture, Sports, Science and Technology.
Author information
Authors and Affiliations
Contributions
Molecular analysis was performed by M.K., Y.S., M.I., F.K., M.O. and S.Y., placental sample collection and preparation by H.K., M.N., Y.T., K.M. and K. Ko., placental pathological examination by K.M., and blood sampling and phenotype assessment by G.N., T.T., M.N., Y.T., K.M., T.U., H.K., Y.K., H.O., K. Ku. and T.O. The study was designed and coordinated by F.I. and T.O. with later input from A.C.F.-S., and the results were interpreted and discussed by A.C.F.-S., F.I. and T.O. The paper was written by T.O.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5, Supplementary Figures 1–3 (PDF 1027 kb)
Rights and permissions
About this article
Cite this article
Kagami, M., Sekita, Y., Nishimura, G. et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 40, 237–242 (2008). https://doi.org/10.1038/ng.2007.56
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2007.56
This article is cited by
-
A Comprehensive Review of Syndromic Forms of Obesity: Genetic Etiology, Clinical Features and Molecular Diagnosis
Current Obesity Reports (2024)
-
Circulating miRNA profiles in COVID-19 patients and meta-analysis: implications for disease progression and prognosis
Scientific Reports (2023)
-
Paternal UPD14 with sSMC derived from chromosome 14 in Kagami–Ogata syndrome
Chromosome Research (2023)
-
Diagnostic implications of pitfalls in causal variant identification based on 4577 molecularly characterized families
Nature Communications (2023)
-
Association of Keap1 (rs11085735) polymorphism and lncRNA MEG3 hypermethylation status with the risk of preeclampsia
Egyptian Journal of Medical Human Genetics (2022)