Abstract
The kinetochore is the crucial apparatus regulating chromosome segregation in mitosis and meiosis. Particularly in meiosis I, unlike in mitosis, sister kinetochores are captured by microtubules emanating from the same spindle pole (mono-orientation) and centromeric cohesion mediated by cohesin is protected in the following anaphase. Although meiotic kinetochore factors have been identified only in budding and fission yeasts, these molecules and their functions are thought to have diverged earlier. Therefore, a conserved mechanism for meiotic kinetochore regulation remains elusive. Here we have identified in mouse a meiosis-specific kinetochore factor that we termed MEIKIN, which functions in meiosis I but not in meiosis II or mitosis. MEIKIN plays a crucial role in both mono-orientation and centromeric cohesion protection, partly by stabilizing the localization of the cohesin protector shugoshin. These functions are mediated mainly by the activity of Polo-like kinase PLK1, which is enriched to kinetochores in a MEIKIN-dependent manner. Our integrative analysis indicates that the long-awaited key regulator of meiotic kinetochore function is Meikin, which is conserved from yeasts to humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Uhlmann, F., Lottspeich, F. & Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37–42 (1999)
Peters, J. M., Tedeschi, A. & Schmitz, J. The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 (2008)
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 (2009)
Buonomo, S. B. et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398 (2000)
Kitajima, T. S., Miyazaki, Y., Yamamoto, M. & Watanabe, Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 22, 5643–5653 (2003)
Tachibana-Konwalski, K. et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 24, 2505–2516 (2010)
Moore, D. P. & Orr-Weaver, T. L. Chromosome segregation during meiosis: building an unambivalent bivalent. Curr. Top. Dev. Biol. 37, 263–299 (1997)
Brar, G. A. & Amon, A. Emerging roles for centromeres in meiosis I chromosome segregation. Nature Rev. Genet. 9, 899–910 (2008)
Watanabe, Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nature Rev. Mol. Cell Biol. 13, 370–382 (2012)
Kitajima, T. S., Kawashima, S. A. & Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517 (2004)
Lee, J. et al. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nature Cell Biol. 10, 42–52 (2008)
Llano, E. et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 22, 2400–2413 (2008)
Kitajima, T. S. et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441, 46–52 (2006)
Riedel, C. G. et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441, 53–61 (2006)
Marston, A. L., Tham, W. H., Shah, H. & Amon, A. A genome-wide screen identifies genes required for centromeric cohesion. Science 303, 1367–1370 (2004)
Katis, V. L. et al. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell 18, 397–409 (2010)
Ishiguro, T., Tanaka, K., Sakuno, T. & Watanabe, Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nature Cell Biol. 12, 500–506 (2010)
Hugerat, Y. & Simchen, G. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics 135, 297–308 (1993)
Klapholz, S. & Esposito, R. E. Recombination and chromosome segregation during the single division meiosis in SPO12–1 and SPO13–1 diploids. Genetics 96, 589–611 (1980)
Tóth, A. et al. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103, 1155–1168 (2000)
Katis, V. L. et al. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr. Biol. 14, 2183–2196 (2004)
Lee, B. H., Kiburz, B. M. & Amon, A. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr. Biol. 14, 2168–2182 (2004)
Yokobayashi, S. & Watanabe, Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123, 803–817 (2005)
Tanaka, K., Chang, H. L., Kagami, A. & Watanabe, Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343 (2009)
Kitajima, T. S., Ohsugi, M. & Ellenberg, J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell 146, 568–581 (2011)
Shonn, M. A., McCarroll, R. & Murray, A. W. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16, 1659–1671 (2002)
Woods, L. M. et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145, 1395–1406 (1999)
Lénárt, P. et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17, 304–315 (2007)
Elia, A. E. et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95 (2003)
Krapp, A., Del Rosario, E. C. & Simanis, V. The role of Schizosaccharomyces pombe dma1 in spore formation during meiosis. J. Cell Sci. 123, 3284–3293 (2010)
Matos, J. et al. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135, 662–678 (2008)
Clyne, R. K. et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nature Cell Biol. 5, 480–485 (2003)
Attner, M. A., Miller, M. P., Ee, L. S., Elkin, S. K. & Amon, A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl Acad. Sci. USA 110, 14278–14283 (2013)
Lee, B. H., Amon, A. & Prinz, S. Spo13 regulates cohesin cleavage. Genes Dev. 16, 1672–1681 (2002)
Monje-Casas, F., Prabhu, V. R., Lee, B. H., Boselli, M. & Amon, A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128, 477–490 (2007)
Corbett, K. D. et al. The monopolin complex crosslinks kinetochore components to regulate chromosome–microtubule attachments. Cell 142, 556–567 (2010)
Sarangapani, K. K. et al. Sister kinetochores are mechanically fused during meiosis I in yeast. Science 346, 248–251 (2014)
Gregan, J. et al. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol. 17, 1190–1200 (2007)
Tada, K., Susumu, H., Sakuno, T. & Watanabe, Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474, 477–483 (2011)
Sakuno, T., Tada, K. & Watanabe, Y. Kinetochore geometry defined by cohesion within the centromere. Nature 458, 852–858 (2009)
Tachibana-Konwalski, K. et al. Spindle assembly checkpoint of oocytes depends on a kinetochore structure determined by cohesin in meiosis I. Curr. Biol. 23, 2534–2539 (2013)
Kagami, A. et al. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 12, 1189–1195 (2011)
Ahonen, L. J. et al. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 15, 1078–1089 (2005)
Kang, Y. H. et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1–PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell 24, 409–422 (2006)
Hodges, C. A., Revenkova, E., Jessberger, R., Hassold, T. J. & Hunt, P. A. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nature Genet. 37, 1351–1355 (2005)
Lister, L. M. et al. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 20, 1511–1521 (2010)
Chiang, T., Duncan, F. E., Schindler, K., Schultz, R. M. & Lampson, M. A. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 20, 1522–1528 (2010)
Jessberger, R. Age-related aneuploidy through cohesion exhaustion. EMBO Rep. 13, 539–546 (2012)
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Rev. Genet. 2, 280–291 (2001)
Nagaoka, S. I., Hodges, C. A., Albertini, D. F. & Hunt, P. A. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol. 21, 651–657 (2011)
Baker, S. M. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet. 13, 336–342 (1996)
Kim-Kaneyama, J. R. et al. Hic-5 deficiency enhances mechanosensitive apoptosis and modulates vascular remodeling. J. Mol. Cell. Cardiol. 50, 77–86 (2011)
Ishiguro, K., Kim, J., Fujiyama-Nakamura, S., Kato, S. & Watanabe, Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 12, 267–275 (2011)
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K. & Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2010)
Morimoto, A. et al. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198, 165–172 (2012)
Chambon, J. P., Hached, K. & Wassmann, K. Chromosome spreads with centromere staining in mouse oocytes. Methods Mol. Biol. 957, 203–212 (2013)
Hodges, C. A. & Hunt, P. A. Simultaneous analysis of chromosomes and chromosome-associated proteins in mammalian oocytes and embryos. Chromosoma 111, 165–169 (2002)
Shibuya, H., Ishiguro, K. & Watanabe, Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nature Cell Biol. 16, 145–156 (2014)
Rabut, G. & Ellenberg, J. Automatic real-time three-dimensional cell tracking by fluorescence microscopy. J. Microsc. 216, 131–137 (2004)
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998)
Sakuno, T., Tanaka, K., Hauf, S. & Watanabe, Y. Repositioning of Aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev. Cell 21, 534–545 (2011)
Acknowledgements
We thank K. Tachibana-Konwalski for the C57BL/6 Mlh1tm1Liskay knockout mice and and J. Ellenberg for a macro for automated microscopy. We also thank J. Lee, T. Hirano, S. Fujiyama, Y. Yamazumi, as well as the Gotoh laboratory for technical advice and all members of the Watanabe laboratory for their support and discussion. This work was supported in part by a JSPS Research Fellowship (to J.K.), SAF2011-25252 (to A.M.P.), a research grant from Uehara Memorial Foundation, a RIKEN CDB intramural grant and a Grant-in-Aid for Young Scientists (B) (to T.S.K.), a Grant-in-Aid for Scientific Research on Innovative Areas, a Grant-in-Aid for Scientific Research (C) (to K.I.), and a Grant-in-Aid for Specially Promoted Research (to Y.W.) from MEXT, Japan.
Author information
Authors and Affiliations
Contributions
J.K., supported by K.I. performed most of the experiments in mice. K.I. and N.T. generated Meikin knockout mice. A.M.P. provided Sgo2 knockout mice. A.N. isolated MEIKIN in yeast two-hybrid screening. B.A., S.Y., A.K., T.I. and T.S. performed experiments in fission yeast. Y.S. and T.S.K. performed live imaging. The experimental design and interpretation of data were conducted by J.K., K.I., B.A., S.Y., A.K., T.I., T.S.K., Y.T. and T.S. Y.W. supervised the project, and wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 MEIKIN (4930404A10rik) was identified as a meiosis-specific CENP-C binding protein.
a, Yeast two-hybrid screening was performed using CENP-C C terminus (amino acids 692–906) as bait and a mouse testis cDNA library as prey. A total of 11.95 × 107 colonies were screened on selective (SD-Trp-Leu-His-Ade, +10 mM 3AT) plates using the AH109 tester strain. The number of clones isolated by screening is summarized. b, To search for a meiosis-specific candidate from the isolated clones, tissue-specific expression patterns of CENP-C interactors were examined by RT–PCR. RNA was extracted from each tissue of both males and females. Testis RNA was derived from 8-week-old males. Ovary RNA was derived from 4- and 8-week-old females. RT(−) indicates control PCR without reverse transcription. Note that the expression of 4930404A10rik is restricted to testis and ovary, as is that of SMC1β. c, Immunoprecipitates from mouse testis chromatin extracts using anti-CENP-C antibody or control IgG were analysed by the indicated antibody. d, The C-terminal domain of MEIKIN (4930404A10rik) (amino acids 385–434) interacts with the CENP-C C terminus in yeast two-hybrid assay.
Extended Data Figure 2 Sequence alignment of MEIKIN homologues in vertebrates.
Amino acid sequences of M. musculus 4930404A10Rik (NP_083381), R. norvegicus (XP_573090), C. lupus (XP_003639413), X. tropicalis (XP_002934413) and G. gallus (XP_001234011) are derived from the NCBI protein database. H. sapiens data are derived from cDNA clones. 4930404A10Rik protein is conserved among vertebrates but it does not have any known motif except for the polo-box binding motif (blue line) in mammalian proteins. Two-hybrid assays indicate that this motif of mouse MEIKIN is important for PLK1 binding (data not shown), although the motif is apparently not conserved in Xenopus and chicken. The C-terminal sequences (red lines) are required for the kinetochore localization (see Fig. 1c).
Extended Data Figure 3 Co-localization of CENP-C and ACA in spermatocytes and MEKIN localization in oocytes.
a, Squashed spermatocytes from wild-type were immunostained for CENP-C, ACA, SYCP3 and DAPI at the indicated stages during meiosis. CENP-C and ACA signals accumulate and co-localize at centromeres after zygotene throughout meiosis. b, Chromosome spreads of oocytes from wild-type were immunostained for MEIKIN, ACA and REC8 at different meiotic stages. Zygotene from E15.5, pachytene and diplotene from E18.5 mice. c, Chromosome spreads of oocytes at metaphase I (5 h after GVBD) and metaphase II (16 h post GVBD) were stained for MEIKIN, ACA and DAPI. Scale bars, 5 μm.
Extended Data Figure 4 Generation of Meikin-knockout mice.
a, Schematic illustrations of the wild-type allele and targeted Meikin−/− allele are shown. Grey boxes represent exons. The targeted exon 4 contains the intact splicing acceptor (SA) sequence followed by a premature stop codon, resulting in disruption of the Meikin allele. Black bar probe for Southern blot. b, Southern blot of genomic DNA from wild type (+/+) and Meikin heterozygous (+/−) ES cells after Pvu II digestion. ES cell clone number 2 used to generate the mice. c, Immunoblot analysis of testis extracts prepared from mice with the indicated genotypes (4-week-old). In Meikin−/−, the specific bands probed by the anti-MEIKIN antibody are absent (shown as arrowheads). α-tubulin is a loading control. d, Testes from 12-week-old wild-type (+/+) and Meikin−/− (−/−) mice. e, Haematoxylin and eosin staining of a section of the testis (12-week-old) showed seminiferous tubules. Enlarged pictures of seminiferous tubules showed spermatocyte (black arrowheads) and spermatids (yellow arrowheads) in wild-type and Meikin−/−. Scale bar, 100 μm. f, Haematoxylin-eosin staining of epididymis from 12-week-old mice shows reduced number of sperms in Meikin−/ −. Enlarged images of sperms are shown. Scale bar, 50 μm. g, A pair of ovaries (8-week-old) from the indicated genotypes (top). Haematoxylin and eosin–stained paraffin sections of ovaries from 8-week-old wild-type and Meikin−/− mice (middle). The antral-stage follicles with oocyte nuclei are magnified (bottom). Asterisks indicate corpora lutea. Scale bar, 500 μm.
Extended Data Figure 5 The delay of anaphase I onset in Meikin−/− oocytes is suppressed by inactivation of SAC.
Oocytes from wild-type and Meikin−/− mice were cultured after GVBD in the presence of nocodazole (10 μM) or the Mps1 kinase inhibitor reversin (5 μM). The first polar body extrusion (PBE) rates are shown with mean ± s.e.m. from 3 independent experiments. The total number of oocytes is n = 27 in wild-type with nocodazole (7, 10 and 10, respectively), n = 27 for wild-type with reversine (6, 10 and 11. respectively), n = 21 for Meikin−/− with nocodazole (5, 8 and 8, respectively) and n = 23 for Meikin−/− with reversine (7, 8 and 8, respectively).
Extended Data Figure 6 MEIKIN is required for centromeric cohesion protection and mono-orientation in spermatocytes.
a, Squashed spermatocytes at metaphase I (Meta I) were immunostained for SGO2, ACA and DAPI. Partial z-axis projection images of aligned chromosomes are shown with magnified images of a bivalent (top). The signal intensity of SGO2 adjacent to the centromere was quantified and normalized to that of ACA. The relative intensities are shown with mean + s.e.m. from 3 independent experiments (bottom). In each experiment, 15 centromeres from a spermatocyte were quantified (n = 4 cells). b, Squashed spermatocytes at anaphase I (5 μm < segregated DNA mass distance <10 μm) from wild-type, Meikin−/− and Sgo2−/− mice were immunostained for CENP-C, REC8 and DNA (top). A pair of sister kinetochores is magnified. The distance between sister kinetochores was scored and represented in the scatter plot with median (bottom). A total of 15 kinetochores from 5 spermatocytes were measured in each group (n = kinetochore number). c, Squashed spermatocytes at prometaphase I from wild-type and Meikin−/− mice, and MEF cells at prometaphase (prometa) were immunostained for CENP-C and DNA. Pairs of sister kinetochores are magnified. The distances between sister kinetochores were scored and represented in the graph with mean + s.e.m. from three independent experiments (right). Note that, MEF cells sample preparation and sister kinetochore distance measurements were performed same methods with spermatocytes. In each experiment, 10 kinetochores were measured in a cell (n = 5 cells). *P < 0.05, *** P < 0.001, **** P < 0.0001, unpaired t-test (a–c). Scale bars, 5 μm (unless otherwise indicated).
Extended Data Figure 7 PLK1 was identified as a MEIKIN interactor.
a, Yeast two-hybrid screening of mouse MEIKIN interactors was performed using a mouse testis cDNA library. The number of clones isolated by screening is summarized. Because the use of MEIKIN full-length and N-terminal region (amino acids 1–271) as bait resulted in a high background of false positive interactions in yeast two-hybrid screening, we used MEIKIN C terminus (amino acids 272–434) as bait. b, Yeast two-hybrid assay demonstrates that MEIKIN interacts directly with mouse PLK1 through the MEIKIN C-terminal domain (amino acids 272–434). c, Mouse MEIKIN protein was immunoprecipitated from testis chromatin-bound fraction by two different anti-MEIKIN polyclonal antibodies (A and B). The immunoprecipitates underwent two independent LC–MS/MS analyses. Those proteins commonly identified in all the LC–MS/MS analyses are listed with the number of peptide hits in the table. Note that polo-like kinase (PLK1) as well as CENP-C were repeatedly identified in the two-hybrid screening (a) and LC–MS/MS analyses (c).
Extended Data Figure 8 BI 2536 treatment reduces PLK1 kinase activity in oocytes.
a, Schematic illustration of BI 2536 treatment in wild-type oocyte culture (left). Oocytes were treated with DMSO or BI 2536 (100 nM) during the indicated time periods, then washed and released into normal culture medium. The first polar body extrusion ratio (1st PBE) was counted at 10 h after GVBD (right). Error bars, mean ± s.e.m. from 3 independent experiments. The total number of oocytes used for each experiment is shown. b, Wild-type oocytes treated with DMSO or BI 2536 (during 6–7 h after GVBD) were fixed and immunostained for PLK1 substrate pCENP-U, ACA and DAPI at metaphase I (top). The relative pCENP-U intensity normalized to that of ACA is shown in the graph with mean + s.e.m. from 3 independent experiments. In each experiment, 10 kinetochores from an oocyte were quantified (n = 4 cells). **P < 0.01, unpaired t-test. c, Oocytes were treated with DMSO or BI 2536 during the period of 4–6 h after GVBD and stained for α-tubulin, CENP-C and DAPI (DNA) at the indicated stages in whole mount (related to Fig. 4d). Magnified images are shown to highlight the separation of sister kinetochores in BI 2536-treated oocyte in anaphase I. d, Chromosome spreads from control and BI 2536-treated wild-type oocytes at metaphase I (6 h after GVBD) and metaphase II (20 h after GVBD) were stained for REC8, CENP-C and DAPI (DNA) (related to Fig. 4d). Magnified images are shown to highlight the loss of cohesion in BI 2536 treated oocytes in metaphase II. e, Mlh1−/− oocytes (C57BL/6 background) were treated with BI 2536 between 9–10 h after GVBD and stained for α-tubulin, CENP-C and DAPI (DNA) in whole mount (related to Fig. 4e). Magnified images are shown to highlight bi-oriented sister kinetochores at the spindle midzone. Scale bars, 5 μm (unless otherwise indicated).
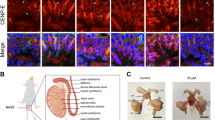
Extended Data Figure 9 The human homologue of MEIKIN.
a, Schematic illustrations of mouse and human MEIKINs. The putative amino acid sequence of hMEIKIN full-length (373 amino acids) was deduced from our own sequencing of DNA (see Extended Data Fig. 2), which was amplified by RT–PCR from the human testis cDNA. The amino acid sequence equivalent to mouse exon 10 (mEX10) is absent in the hMEIKIN protein, despite our attempts at a computational search to identify the missing DNA sequence. b, Yeast two-hybrid assays demonstrate that the hMEIKIN C terminus interacts with human CENP-C (full length), hCENP-C C-terminal (CENPC motif + Mif2 motif, amino acids 732–945) and hPLK1, in agreement with mouse MEIKIN data. c, Immunostaining of a human seminiferous tubule section (purchased from Biochain) demonstrates that hMEIKIN localizes to centromeres (ACA) in pachytene spermatocytes. We used anti-hMEIKIN-N and anti-hMEIKIN-C antibodies. Enlarged images of the rectangles are shown to highlight the co-localization of ACA and hMEIKIN (bottom). Scale bar, 5 μm.
Supplementary information
Supplementary Data
This file contains Supplementary Table 1. (PDF 96 kb)
: Live imaging of wild-type (WT) and Meikin -/- (KO) oocytes expressing 2mEGFP-CENP-C (green) and H2B-mCherry (red) from metaphase I to anaphase I
Kinetochore signals are peak-enhanced and background-subtracted. Time after the end of metaphase I (min). Scale bar, 10 μm. (AVI 29704 kb)
Live imaging of wild-type (WT) and Meikin -/- (KO) oocytes expressing 2mEGFP-CENP-C (green) and H2B-mCherry (red) from telophase I to metaphase II
Kinetochore signals are peak-enhanced and background-subtracted. Time after telophase I (hh:mm). Scale bar, 10μm. (AVI 25823 kb)
Rights and permissions
About this article
Cite this article
Kim, J., Ishiguro, Ki., Nambu, A. et al. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517, 466–471 (2015). https://doi.org/10.1038/nature14097
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14097
This article is cited by
-
Distinct dynein complexes defined by DYNLRB1 and DYNLRB2 regulate mitotic and male meiotic spindle bipolarity
Nature Communications (2023)
-
Engineering apomixis in crops
Theoretical and Applied Genetics (2023)
-
Asynapsis and unreduced gamete formation in a Trifolium interspecific hybrid
BMC Plant Biology (2022)
-
Genome-wide DNA methylation dynamics during epigenetic reprogramming in the porcine germline
Clinical Epigenetics (2021)
-
Meiosis-specific ZFP541 repressor complex promotes developmental progression of meiotic prophase towards completion during mouse spermatogenesis
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.