Abstract
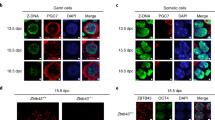
Mammalian genomes employ heritable cytosine methylation in the long-term silencing of retrotransposons and genes subject to genomic imprinting and X chromosome inactivation. Little is known of the mechanisms that direct cytosine methylation to specific sequences. Here we show that DNA methyltransferase 3-like (Dnmt3L (ref. 1)) is expressed in testes during a brief perinatal period in the non-dividing precursors of spermatogonial stem cells at a stage where retrotransposons undergo de novo methylation. Deletion of the Dnmt3L gene prevented the de novo methylation of both long-terminal-repeat (LTR) and non-LTR retrotransposons, which were transcribed at high levels in spermatogonia and spermatocytes. Loss of Dnmt3L from early germ cells also caused meiotic failure in spermatocytes, which do not express Dnmt3L. Whereas dispersed repeated sequences were demethylated in mutant germ cells, tandem repeats in pericentric regions were methylated normally. This result indicates that the Dnmt3L protein might have a function in the de novo methylation of dispersed repeated sequences in a premeiotic genome scanning process that occurs in male germ cells at about the time of birth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Aapola, U. et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65, 293–298 (2000)
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002)
Bourc'his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001)
La Salle, S. et al. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 268, 403–415 (2004)
Dobson, M. J., Pearlman, R. E., Karaiskakis, A., Spyropoulos, B. & Moens, P. B. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 107, 2749–2760 (1994)
Odorisio, T., Rodriguez, T. A., Evans, E. P., Clarke, A. R. & Burgoyne, P. S. Meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nature Genet. 18, 257–261 (1998)
Enders, G. C. & May, J. J. Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev. Biol. 163, 331–340 (1994)
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997)
Lin, S. P. et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1–Gtl2 imprinted cluster on mouse chromosome 12. Nature Genet. 35, 97–102 (2003)
Davis, T. L., Trasler, J. M., Moss, S. B., Yang, G. J. & Bartolomei, M. S. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58, 18–28 (1999)
Ueda, T. et al. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5, 649–659 (2000)
Bestor, T. H. Cytosine methylation mediates sexual conflict. Trends Genet. 19, 185–190 (2003)
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 20, 116–117 (1998)
Kuff, E. L. & Lueders, K. K. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51, 183–276 (1988)
Ostertag, E. M. & Kazazian, H. H. Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538 (2001)
Kaneda, M. et al. Essential role for de novo methyltransferase 3a in paternal and maternal imprinting. Nature 429, 900–903 (2004)
Bestor, T. H. & Tycko, B. Creation of genomic methylation patterns. Nature Genet. 12, 363–367 (1996)
Chedin, F., Lieber, M. R. & Hsieh, C. L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA 99, 16916–16921 (2002)
Cambareri, E. B., Jensen, B. C., Schabtach, E. & Selker, E. U. Repeat-induced G-C to A-T mutations in Neurospora. Science 244, 1571–1575 (1989)
Maloisel, L. & Rossignol, J.-L. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 12, 1381–1389 (1998)
Petes, T. D. Meiotic recombination hot spots and cold spots. Nature Rev. Genet. 2, 360–369 (2001)
Peters, A. H., Plug, A. W., van Vugt, M. J. & de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997)
Ponzetto-Zimmerman, C. & Wolgemuth, D. J. Methylation of satellite sequences in mouse spermatogenic and somatic DNAs. Nucleic Acids Res. 12, 2807–2822 (1984)
Sanford, J., Forrester, L. & Chapman, V. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 12, 2822–2835 (1984)
Pietras, D. F. et al. Construction of a small Mus musculus repetitive DNA library: identification of a new satellite sequence in Mus musculus. Nucleic Acids Res. 11, 6965–6983 (1983)
Acknowledgements
We thank B. Tycko for suggestions, P. Moens for antibodies against synaptonemal complex proteins and H1t and for discussions, G. Enders for antibodies against GCNA1, J. Walonoski for advice on in situ hybridization, and K. Anderson, M. Damelin and M. Goll for criticism of the manuscript. This work was supported by grants from the National Institutes of Health to T.H.B. and by a fellowship from the Rett Syndrome Research Foundation to D.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Bourc'his, D., Bestor, T. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). https://doi.org/10.1038/nature02886
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02886
This article is cited by
-
DNMT1 SNPs (rs2114724 and rs2228611) associated with positive symptoms in Chinese patients with schizophrenia
Annals of General Psychiatry (2023)
-
Expression analysis suggests that DNMT3L is required for oocyte de novo DNA methylation only in Muridae and Cricetidae rodents
Epigenetics & Chromatin (2023)
-
The interferon stimulated gene-encoded protein HELZ2 inhibits human LINE-1 retrotransposition and LINE-1 RNA-mediated type I interferon induction
Nature Communications (2023)
-
Emerging evidence that the mammalian sperm epigenome serves as a template for embryo development
Nature Communications (2023)
-
Nuclear cGAS restricts L1 retrotransposition by promoting TRIM41-mediated ORF2p ubiquitination and degradation
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.