Abstract
Nuclear pore complexes (NPCs) are large channels spanning the nuclear envelope that mediate nucleocytoplasmic transport. They are composed of multiple copies of ~30 proteins termed nucleoporins (NUPs). Alterations in NUP genes are linked to several human neoplastic and non-neoplastic diseases. This review focuses on NUPs, their genes, localization, function in the NPC and involvement in human diseases.
Similar content being viewed by others
Introduction: NPC structure and nucleoporins
At the fusion point of the inner and outer nuclear envelope (NE) membranes, ~600 copies of ~30 proteins termed Nucleoporins (NUPs) assemble to form the nuclear pore complex (NPC). The NPC, one of the largest macromolecules in the cell, forms an octagonal channel across the NE, which serves to mediate nucleocytoplasmic transport.1 The number of NPCs per nucleus varies with the species, environmental conditions, cell activities and the cell cycle as it doubles during interphase and before mitosis and reaches an apex during the S-phase.2
The symmetrical NPC core can be viewed as a series of concentric cylinders: an outer ‘pore membrane’ (orange cylinder), a ‘coat’ (blue cylinder), an ‘adaptor’ (yellow cylinder) and an inner ‘transport’ channel (pink cylinder) surrounding a central channel with a diameter of 40–50 nm (Figure 1).3 Sixteen filaments stretch from the NPC core to the cytoplasm (brown filaments, Figure 1) and nucleoplasm (violet filaments, Figure 1). A distal ring links the eight filaments in the nucleoplasm so they form a basket-like structure3 (Figure 1).
Although the majority of NUPs are followed by a number that, in most cases, refers to their molecular mass, a uniform NUP nomenclature still does not exist3 since the molecular mass of each one varies between species.2 According to their approximate localization within the NPC core or filaments, NUPs fall into six main categories:3 (I) transmembrane NUPs (Figure 1, orange cylinder), NUP210, NDC1 and POM121, are integral membrane proteins, bearing single (ie, NUP210 and POM121) or multiple (ie, NDC1) transmembrane domains. They are thought to have a role in NPC assembly and anchoring to the NE.3 (II) Membrane-apposed coat NUPs (Figure 1, blue cylinder) include NUP160, NUP133, NUP107, NUP96, NUP85, SEH1L, NUP43, NUP37 and SEC13. They form the nonameric Nup107–160 complex which, in size and complexity, is the largest NPC subunit and is required for early steps in NPC assembly. An additional protein, ELYS, is thought to be the tenth member of the Nup107–160 complex.3 (III) Adapter NUPs (Figure 1, yellow cylinder), NUP205, NUP188, NUP155, NUP93 and NUP35, constitute the NUP93 complex, which is the second largest NPC structural unit. NUP188 and NUP93 have a crucial role in establishing the NE subcompartments, such as the outer nuclear membrane (ONM) that is continuous with the endoplasmic reticulum and the inner nuclear membrane (INM). Altogether, NUP188 and NUP93 form a barrier that prevents membrane proteins from passing from the ONM to the INM. NUP155 ensures correct INM protein localization.4 (IV) Channel NUPs (Figure 1, pink cylinder), that is, NUP62, NUP58 (alias NUPL1) and its splice variants NUP45 and NUP54, constitute the innermost cylindrical layer of the NPC. Although the precise channel structure and composition is still unknown, NUP54/NUP58 forms a midplane ring that undergoes large-scale, reversible expansion to regulate nucleocytoplasmic traffic.5 (V) Nuclear basket NUPs (Figure 1, violet filaments), that is, NUP153, TPR and NUP50 constitute NPC cytoplasmic filaments, being involved in nuclear import/export processes.1 In particular, NUP153 and NUP50 form a ring that binds TPR protein filaments, making up a basket that facilitates nucleocytoplasmic transport.3 (VI) NPC cytoplasmic filaments (Figure 1, brown filaments) are composed of seven proteins, RANBP2 (alias NUP358), NUP214, NUP98, NUP88, ALADIN, NUPL2 (alias hCG1) and RAE1, providing key interaction sites for nucleocytoplasmic transport machinery.3 Two other proteins involved in mRNA trafficking are mRNA export factor GLE1 and the DEAD-box-containing RNA helicase DDX19, which are constitutively attached to cytoplasmic filaments and are considered as part of the NPC.3
It is worth noting that some NUPs (ie, NUP98, NUP62, NUP50, NUP153) are not static components of the pore but instead move on and off of the NPC, shuttle in and out of the nucleus, and associate with cytoplasmic and nuclear structure.
Independent of location approximately one-third of NUPs contain phenylalanine–glycine (FG) repeats. FG repeats fill the central transport channel and, as they lack an ordered secondary structure, can adopt different conformations and associate with cargo transport receptors to act as a barrier to non-specific cargo transport.1
Controlling nucleocytoplasmic transport is the main NPC function. Ions and small molecules diffuse freely through the pore while larger molecules, over 40–60 kDa in size, need to be shuttled. Shuttling requires nuclear transport receptors6 and specific transport signals, that is, a nuclear localization signal (NLS) or a nuclear export signal (NES). Soluble transport receptor proteins of the karyopherin family (known as importins and exportins) recognize NLSs or NESs either directly or through adapter proteins and karyopherin macromolecule binding mediates translocation.7 Although nucleocytoplasmic transport is governed by several types of protein–protein interactions, GTP hydrolysis by the Ran GTPase is the only enzymatic reaction to occur. RanGTP binding to import complexes causes their dissociation, while Ran GTPase-activating protein stimulated hydrolysis in the cytoplasm triggers the disassembly of RanGTP–karyopherin macromolecule complexes.6, 7
Besides enabling nucleocytoplasmic transport, NUPs are involved in many fundamental cellular processes such as differentiation, gene expression, chromatin organization, epigenetic regulation, replication-coupled DNA repair and mitosis.3, 6, 8 Consequently, it is hardly surprising that genetic alterations in many NUP genes are linked to cellular and developmental defects, as well as to various human diseases, including autoimmune dysfunctions, neurological diseases, cardiovascular disorders and cancer.3, 9 Table 1 summarizes each group of NUPs, their genetic abnormalities and their involvement in human neoplastic and non-neoplastic disorders.
NUPs and diseases
Immune diseases
In several autoimmune diseases anti-NPC autoantibodies recognize diverse NUPs. As significant similarities were noticed between NUPs and glycoproteins in many animal viruses, NPC autoantibody generation in autoimmune diseases is hypothesized to be due to molecular mimicry.10, 11
In primary biliary cirrhosis (PBC), bile ducts deteriorate gradually, resulting in the development of liver cirrhosis. It is still unclear whether the autoimmune assault leading to cell destruction, loss of immunological tolerance and immune response induction against NPC antigens is the primary cause of PBC or whether it is an independent epiphenomenon.11 Anti-NUP210 and –NUP62 antibodies were found in 9.4–41.2% and 22–32% of patients with PBC, respectively.11 Although anti-NUP210 antibodies had a specificity of ≥96% and were thought to predict poor outcome, the prognostic significance and specificity of anti-NUP62 antibodies remains to be established.11
In other autoimmune disorders, such as mixed connective tissue diseases, rheumatic diseases, systemic lupus erythematosus and Sjögren’s syndrome, reactivity against NUP210, NUP62, RANBP2, NUP153 and TPR was found sporadically10, 11 (Table 1).
Both NUP93 and RANBP2 mediate nucleocytoplasmic transport of AIRE, a crucial transcriptional regulator that acts on gene expression and influences clonal deletion of differentiating T cells in the thymus. In thymuses from patients with Down syndrome (DS) downregulation of both AIRE and NUP93 was found linking global thymic hypofunction observed in DS to the deregulated expression of an NPC protein.12
Upregulation of NUP54 gene has been recently found in psoriatic T cells that mediate the chronic inflammation typical of psoriasis.13
Anecdotal evidence from in vitro and in vivo models suggests the Nup107–160 complex members are involved in immune dysfunctions. Nup96(+/−) mice showed impaired antigen presentation and T-cell proliferation as well as a high susceptibility to viral infection, suggesting that Nup96 has a role in innate and adaptive immunity.14 During inflammation in mice and human cell lines, Nup85 bind chemokine receptors mediating leukocyte and monocyte migration.15
Viral infections
Many human viruses target NUPs and alter the NPC structure, composition and function.9 Viruses are generally grouped into two categories, those with DNA genomes and those with RNA genomes.2 Mechanisms underlying pathogenesis vary greatly and each virus deals with the NPC in a distinct way.2
DNA viruses
After fusing their viral envelope with the plasma membrane, DNA viruses enter the cytoplasm and pass through the NPC. In the nucleus they fuse their genomes with the host cell genome to proliferate.2
Small viruses, such as the hepatitis B virus (HBV), travel easily through the NPC. After capsid core protein phosphorylation exposes a C-terminal NLS, the NLS is more easily recognized by importin-α and importin-β to facilitate translocation.16 The import complex then interacts with NUP153 in the nuclear basket by binding to FxFG repeats.17
Larger viruses, like the herpes simplex viruses (HSV) and adenoviruses need to disassemble from the viral capsid to access the nucleus for proliferation.2
HSV1, the best characterized HSV for its nuclear import strategy, dissociates only partially from the capsid in the cytoplasm and then binds the NPC. Its behavior is similar to the bacteriophage infection pattern as NPC binding triggers viral DNA injection into pore channels.18 Experimental evidence suggested that specific interactions between viral proteins and RANBP2 bring the capsid and NPC together and promote contacts with NUP214, which ultimately lead to viral genome ejection.19
NUP214 and RANBP2 are also involved in adenovirus interactions with the NPC. The viral capsid binds NUP214 and the motor protein kinesin-1 that is also connected with RANBP2. The movement of kinesin-1 produces a pulling action that disassembles capsid proteins and NUPs. Greater nuclear permeability enables uncoated viral genome entry into the nucleus.19
Several lines of evidence support the hypothesis that polyomaviruses, whose prototype is the simian virus 40 (SV40), may follow either NPC-dependent or –independent nuclear entry pathways. The former mechanism involves importin-α-/β-dependent trafficking through the NPC while the latter requires NE disruption.19 The molecular mechanisms underlying these pathways are still not fully understood.
Direct interaction with the NPC may not be involved in parvovirus and papillomavirus entry into the nucleus as parvoviruses seem to destroy the NE, while papillomaviruses appear to exploit NE breakdown during mitosis.19
RNA viruses
RNA viruses proliferate in the host cell cytoplasm but these viruses may target the NPC to improve viral replication and transmission.2 For example, polioviruses and rhinoviruses induce proteolytic degradation of NUPs, particularly NUP62, NUP153 and NUP98.20
Interestingly, one viral strategy to promote RNA virus replication may be NUP98 and RAE1 downregulation. NUP98 is downregulated in cells infected by influenza A virus, while NUP98 and RAE1 are induced by antiviral cytokines such as interferons.21 Furthermore, the vescicular stomatitis virus (VSV), which preferentially infects cancer cells,9 produces the so-called M protein that dissociates RAE1 from NUP98, thus inhibiting mRNA export processes, cell transcription and mitosis progression.22
HIV-1, the most extensively studied RNA virus, underlies the AIDS, which is caused by infection-related depletion of CD4+ T cells and macrophages.23 HIV-1 attaches to the cell surface and fuses with the cytoplasmic membrane, whereupon the capsid is released into the cytoplasm and traffics toward the nucleus, while the viral RNA genome is reverse-transcribed into DNA.23 RANBP2 and NUP153 appear crucial for HIV-1 docking at the NPC and for its transport inside the nucleus.9 There the viral genome integrates at highly expressed chromosomal locations that directs the formation of progeny virions.24 Among the NUPs associated with transcriptional active chromatin, NUP62, NUP98, NUP153 and TPR, the last two seem to have a role in HIV-1 infection. NUP153 regulates the viral nuclear localization while TPR might interfere with viral replication favoring contacts with hypertranscribed genes.24
Finally, in HIV-1 infected cells, NUP43, NUP45, NUP54 and NUP58/NUPL1 are downregulated while NUP35, NUP98 and TPR are upregulated.9 NUP62 might also have a role in HIV-1 assembly and infectivity as its small interfering RNA (siRNA)-mediated downregulation decreased viral protein synthesis and viral production.9
Neurological diseases
The Triple A syndrome, an autosomal recessive disorder characterized by adrenal failure and abnormal autonomic nervous system development, results from ALADIN gene variants.25 Variant-related mislocalization of the protein from cytoplasmic filaments appears to underlie disease pathogenesis. Fibroblasts from Triple A syndrome patients displayed a selective failure to import DNA repair proteins into the nucleus with accumulating DNA damage leading to cell death.26 ALADIN knockdown produced the same effects in human adrenal cells, which was also associated with an unbalanced redox homeostasis and a downregulation of genes required for steroidogenesis.27
In three motor neuron degenerative diseases, amyotrophic lateral sclerosis (ALS), lethal congenital contracture syndrome type I and lethal arthrogryposis with anterior horn cell disease, deleterious GLE1 variants were reported.28 Independent of protein location at the NPC, which may- or may not- be altered, they all cause defects in mRNA metabolism that impact on motoneuron development and survival.28
RANBP2 variants induce susceptibility to familial and recurrent acute necrotizing encephalopathy, which is usually diagnosed in young children after influenza virus infection and is characterized by multiple, symmetrical lesions in the central nervous system.29
In patients with ALS, irregularities in the distribution of the channel FG-NUP62 were described at the NE in anterior horn cells.30 In addition, an autosomal recessive infantile bilateral striatal necrosis was associated with a NUP62 homozygous variant that did not delocalize the protein, suggesting alternative pathogenetic mechanisms.31
A splice site variation of NUP107 segregated in a family with global developmental delay, light complexion and early onset glomerulosclerosis.32 Notably, an hypothetical NUP107 involvement in kidney development was suggested by biallelic NUP107 variants in nine individuals from five unrelated families with an early onset form of steroid-resistant nephrotic syndrome and by a Nup107 knockdown zebrafish model developing abnormal glomerulus structures.33
In nervous system development, evidence from diverse cell lines and animal models revealed putative roles for NUPs 210, 133 and 107. NUP210 was found to be crucial in human cell line myogenic and neuronal differentiation.34 Nup133 deficiency impaired murine neural lineage differentiation.35 Sec13 depletion abolished early retinal development in zebrafish embryos36 and Nup107 homozygous mutations produced serious developmental defects leading to embryonic death at 5–6 days.37 Finally, Rae1 knockdown in Caenorhabditis elegans impaired axon termination and synapse formation.38
Cardiovascular disorders
Heart failure was associated with changes in the levels and the distribution of specific NUPs (Table 1). Cardiovascular diseases and NUP abnormalities were first clearly linked in a family with atrial fibrillation (AF) segregating with a NUP155 homozygous variant (c.1172G>A, p.R391H, NM_153485), which delocalizes NUP155 from the NPC,39 abolishing its interaction with POM121 and NUP35.40 In mice, low Nup155 levels interrupted Hsp70 mRNA export and Hsp70 protein import, which are crucial factors for cardiac function.39 Nup155 interaction with histone deacetylase 4 (Hdac4) and suppression of hypertrophic cardiomyocyte growth were observed in rats. Most interestingly, a truncated Nup155 mutant suppressed Hdac4-induced gene expression patterns.41 In a patient with Heterotaxy (Htx), a congenital heart disease resulting from abnormalities in left–right body patterning, NUP188 was found in a small 9q34.11 duplication of 148 kb.42 Anomalous left–right body patterning also characterized a Nup188 morpholino knockdown.42 The apparent contradiction of gene duplication/deletion leading to the same phenotype was found by the same authors42 in human Htx with either duplication or deletion of the TGFBR2 gene.
Recent evidences from cell lines and rats suggested the involvement of another adapter NUP, namely NUP35, in cardiac function. In fact NUP35 regulated intracellular cardiomyocyte pH by controlling nucleocytoplasmic trafficking of the mammalian Na+–H+ exchanger-1 (NHE1) mRNA.43 Interestingly, both NUP35 and NHE1 were downregulated in ischemic cardiomyocytes both in vitro and in vivo.43
Cancer
NUP gene fusions
NUPs are directly implicated in cancer in several ways: NUP protein expression levels change, single point variants and chromosomal translocations generating fusion proteins (Table 1).
At least four NUP genes, TPR, RANBP2, NUP214 and NUP98, are termed ‘promiscuous’ as they are found fused to several partner genes to produce diverse oncogenic fusion proteins.
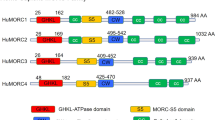
TPR, in its NPC-independent roles, is involved in telomeric chromatin organization, transcription regulation and chromosome segregation during mitosis.9 In its role of proto-oncogene activator, TPR amino-terminal residues fuse, as a consequence of diverse chromosomal translocations, with the protein kinase domains of tyrosine kinase partner genes such as MET (gastric cancer), NTRK1 (thyroid carcinoma), FGFR1 (myeloproliferative syndromes) and ALK (lung adenocarcinoma).9 In all fusion proteins the maintained TPR coiled-coil motif is predicted to allow dimerization that aberrantly activates kinases (Figure 2a and b).
Promiscuous NUP genes. (a) TPR fusion partner genes and their chromosomal localization. (b) In all fusions TPR N-terminal coiled-coil domain is maintained and fused to the partner kinase domain. (c) RANBP2 partner genes and their chromosomal localization. (d) In all fusion proteins RANBP2 N-terminal leucine-rich domain is retained and fused with the tyrosine kinase domain of the partner gene. (e) NUP214 fusion partner genes and their chromosomal localization. (f) Both DEK-NUP214 and SET-NUP214 join DEK or SET protein to the C-terminal two-thirds of NUP214, including a portion of the coiled-coil domain and the FG repeat domain. Similarly NUP214-SQSTM1 chimeric protein fuses N-terminal half of SQSTM1 with a small portion of NUP214 FG repeats domain. Instead NUP214-ABL1 is composed of the N terminus of NUP214, including its β-propeller, coiled-coil domain and FG repeats, fused with most of the ABL1 tyrosine kinase protein. (g) Homeodomain (HD, blue) and non-HD (black) NUP98 fusion partner genes and their chromosomal localization. (h) In all fusion proteins NUP98 preserves GLFG repeats and GLE2-binding domain (GBD), fused with the C-terminal portion of partner genes.
In inflammatory myofibroblastic tumors (IMTs), myeloid malignancies (two adults and three children collected from literature and one child, personal observation) and in diffuse large B-cell lymphoma (one adult), RANBP2 rearranged with tyrosine kinase genes to form RANBP2-ALK fusion (inv(2)(p13q13)/t(2;2)(p23;q13)) (Table 1). In each fusion, the RANBP2 N-terminal leucine-rich domain mediated attachment to the NPC core (Figure 2c and d). In RANBP2-ALK+ cells, ALK delocalization from the cytoplasm to the NE is a useful marker in immunohistochemical diagnosis.44 Interestingly, crizotinib, an oral ALK inhibitor, is being tested in acute myeloid leukemias (AML) and IMT patients with RANBP2-ALK fusion.45 RANBP2-ABL1/t(2;9)(q21;q34) was detected in one pediatric case of high-risk B-cell acute lymphoblastic leukemia (B-ALL); and RANBP2-FGFR1/t(2;8)(q12;p11) was found in an adult with myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN; Figure 2c and d).9, 46
Many reports linked NUP214 to hematological malignancies as it participates in several oncogenic translocations47 (Figure 2e and f), typically associated with a subgroup of MDS/AML in children and adolescents with poor response to therapy is the t(6;9)(p23;q34)/DEK-NUP214. In acute undifferentiated leukemia, AML and T-cell ALL (T-ALL) an interstitial deletion at 9q32–q34 joined SET with NUP214.47 Both SET-NUP214 and DEK-NUP214 are structurally similar, with almost the full SET or DEK proteins joined to two-thirds of the NUP214 C terminus that includes a portion of the coiled-coil domain and the entire FG repeat domain (Figure 2f). SET and DEK are two oncogenes; the first one inhibits cell apoptosis caused by T lymphocytes while the latter is involved in DNA replication and mRNA processing.47 Both SET-NUP214 and DEK-NUP214 fusion proteins localize as punctate ‘dots’ in the nucleoplasm and bind NUP88 and export factors, raising the hypothesis of altered nuclear transport in leukemic cells.48 In SET-NUP214+ T-cell acute leukemia, an activated HOXA gene signature may trigger the leukemogenic process.49 DEK-NUP214 induces in vitro proliferation by upregulation of mTOR pathway.50 Its overexpression increased mRNA translation in myeloid cells due to hyper-phosphorylation of eIF4E, an eukaryotic initiation factor.51 In the final analysis, the leukemogenic function of these fusions is still largely undetermined.
Found in T-ALL, and less frequently in B-ALL,46 a third NUP214 rearrangement, NUP214-ABL1, derives from an ‘in situ’ or an episomal amplification of a small region at chromosome band 9q34. The NUP214 N-terminus (including its β-propeller, coiled-coil and varying amounts of FG repeat regions) fuses to most of the ABL1 protein (Figure 2f). The NUP214-ABL1 fusion proteins localize at the NPC by binding to NUP88. Dimerization and cross-phosphorylation then aberrantly activate the ABL1 kinase domain.
Anecdotal evidence from one case of T-ALL showed NUP214 underwent fusion with SQSTM1, a multifunctional adapter protein, in a cytogenetically cryptic unbalanced der(5)t(5;8)(q35;q34).47 The NUP214-SQSTM1 chimeric protein fused half of the SQSTM1N-terminal with a small portion of NUP214 FG repeats domain (Figure 2f).47
The promiscuous gene NUP98 is well-known for its involvement in hematological malignancies (Table 1). At least 32 different NUP98 partner genes were identified in AML, chronic myeloid leukemia in blast crisis, MDS and T-lymphoid malignancies (Figure 2g).52 NUP98 interacts with the Nup107–160 complex, through direct binding to NUP96, and with the NUP214–NUP88 complex. It anchors to the NPC center through its C-terminal domain and its N-terminal GLFG repeats protrude throughout the NPC being found on both sides of the NPC.53 GLFG repeats, which are thought to function as a docking site during molecule trafficking, are intersected by a coiled-coil domain, the Gle2-binding sequence motif, which mediates binding to RAE1.54 Remarkably, NUP98 is dynamic and mobile, being also found in the nucleoplasm.54 In fact, it is not only involved in nucleocytoplasmic transport but also acts as a transcription factor since GLFG repeats interact with CBP/p300 complex and HDACs.54 NUP98 associates dynamically with the human genome during differentiation, regulates developmental gene expression programs and promotes epigenetic transcriptional memory.55 In fact, a physical association between NUP98 and histone-modifying complexes, that is, MBR-R2/NSL and Trx/MLL, was demonstrated.56
In NUP98 fusions, the NUP98 N-terminal portion fuses with the C-terminal portion of either a homeodomain (HD) or non-HD protein (Figure 2g and h).54 All fusion proteins have the same structure and always involve CD34 antigen-expressing cells or hematopoietic stem cells in myeloid and T-cell leukemias.57
The most frequent NUP98 oncogenic fusion is NUP98-NSD1 as it was found in 16.1% and 2.3% of pediatric and adult AML, respectively, with normal karyotype and poor prognosis.52
Common themes in the leukemogenic properties of NUP98-HOXA9, NUP98-HOXA10, NUP98-HOXD13, NUP98-PMX1, NUP98-HHEX, NUP98-TOP1, NUP98-NSD1 and NUP98-JARID1A, as demonstrated in mice, were anemia and aberrant myeloid lineage differentiation.54 In transgenic mice the Nup98-HD fusion promoted self-renewal and aberrant gene expression to a significantly greater extent than non-HD partners.58 FLT3/ITD and loss of TP53 may favor Nup98 fusion leukemogenic activity.59, 60 NUP98 fusions mediate transcriptional misregulation, which leads to marked HOXA gene overexpression and oncogenic potential.54
As the NUP98 partner gene’s contribution to the fusion’s oncogenic properties has not yet been fully clarified, alternative leukemogenic mechanisms were investigated. Nup98-Ccdc28a induced a rapid, transplantable myeloid neoplasm in recipient mice without Hoxa-Meis1 pathway deregulation.61 NUP98-HOXD13 fusion gene impaired lymphocyte differentiation and non-homologous end-joining-mediated DNA break repair, providing evidence that NUP98 was linked to genomic (in)stability.62 In human and mouse cells bearing NUP98-HOXA9 fusion, the RAE1 NUP was reduced and mislocalized from the mitotic spindle into the nucleus in concomitance with mitotic chromosomal separation defects and enhanced apoptosis.63 Interactions of NUP98 fusion oncoproteins with spindle assembly checkpoint members may cause mitotic spindle defects and chromosome missegregation.64
In murine and human hepatocellular carcinomas (HCC) NUP98 expression was reduced concomitantly with p21 as the 3′UTR of NUP98 was shown to protect p21 mRNA from exosome-mediated degradation.65 This HCC model suggested that NUP98 have a tumor suppressor role through the regulation of a p53 target gene. However, a putative oncogenic effect emerged in ~12% HCC patients that displayed NUP98 overexpression.65 Altogether, these data and the previously discussed results on translocations in hematological malignancies are consistent with a multifaceted role of NUP98 in cancerogenesis.
In B-ALL the PAX5-POM121 fusion protein derives from a leukemogenic translocation between chromosomes 7 and 9.66 It may act as an aberrant transcription factor, as it localizes in the nucleus while maintaining typical endogenous PAX5 binding to its DNA target sequences.67
Anecdotal evidence (one dedifferentiated liposarcoma), showed NUP107/12q15 was fused to the LGR5 gene, located 2.8 Mb distally on 12q.68 Its significance, however, is controversial as it could have been a bystander event in the juxtaposition of multiple amplicons at 12q13–q15 forming a giant chromosome marker, which is the lipoma hallmark.68
NUP gene/protein deregulation
In solid and hematological tumors NUP88 overexpression has been well documented (Table 1). Its underlying molecular mechanism has not yet been well established. Over-expressed NUP88 aggregated in granular dots in the cytoplasm and NUP88 staining intensity correlated with high-grade malignancies, poor differentiation, tumor invasiveness, high proliferation and metastasis.9 Interestingly, NUP88, like NUP214, localizes at the spindles. Altered NUP88 expression enhanced multi-nucleated cells and multipolar spindle formation potentially linking its deregulation with aneuploidy.69 NUP88 depletion by siRNA caused accumulation of NF-κB in the cytoplasm as a consequence of increased nuclear export. As NF-κB is found in the nucleus of many cancer types, a model in which overexpression of NUP88 leads to decreased NF-κB nuclear export70 and upregulation of its target genes has been proposed.9
In solid and hematological tumors other deregulated NUPs include NUP210, NUP133, NUP107, SEC13, NUP188, NUP93, NUP62, NUP153, TPR, RANBP2, NUP214, NUP98, NUPL2 and RAE1 (Table 1).
Finally, although development of several tumors, like, for example, colon and breast cancers, was occasionally linked to sequence variation in NUP genes (Table 1), it does not seem to be the pre-eminent genomic mechanism underlying NUP gene involvement in cancer.
Conclusions
This review has shown that studying NUP pathways has been crucial to understanding their involvement in disease processes, while investigating the role of NUPs in pathologies has led to better understanding of their NPC-dependent and -independent functions. Thus a reciprocal connection is emerging between basic NPC morpho-functional features and NUP disease-related abnormalities. We now know that the NPC is a multifunctional cellular structure with at least 30 NUPs and that specific NUPs and NPC traffic mechanisms are involved in the pathophysiology of cardiac tissue and the immune and nervous systems (Table 1). Many viruses such as HBV,HSV, VSV and HIV-1, use the NPC to access the nucleus and proliferate while recurrent NUP gene abnormalities, particularly in NPC genes encoding for nuclear basket and cytoplasmic filament NUPs, are key events in the development of hematological malignancies as well as solid tumors (Table 1).
Pathways leading from a NUP defect to disease are surely multi-factorial. Pathological events that are unrelated to the NPC probably combine with alterations in nucleocytoplasmic traffic pathways like a reduction in key proteins at the NPC, NUP mistargeting or other transport factors. Therapeutic agents that attempt to normalize protein localization target various regulatory components within the transport process such as cargo proteins, transport receptors, Ran regulators and the NPC itself. Future studies aimed at better understanding the nuclear transport mechanisms, and how they relate to pathogenesis, will probably disclose the identity of novel targets for the treatment of NPC-related diseases.
References
Wälde S, Kehlenbach RH : The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol 2010; 20: 461–469.
Jamali T, Jamali Y, Mehrbod M, Mofrad MR : Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int Rev Cell Mol Biol 2011; 287: 233–286.
Hoelz A, Debler EW, Blobel G : The structure of the nuclear pore complex. Annu Rev Biochem 2011; 80: 613–643.
Busayavalasa K, Chen X, Farrants AK, Wagner N, Sabri N : The Nup155-mediated organisation of inner nuclear membrane proteins is independent of Nup155 anchoring to the metazoan nuclear pore complex. J Cell Sci 2012; 125: 4214–4218.
Solmaz SR, Chauhan R, Blobel G, Melčák I : Molecular architecture of the transport channel of the nuclear pore complex. Cell 2011; 147: 590–602.
Raices M, D'Angelo MA : Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 2012; 13: 687–699.
Cautain B, Hill R, de Pedro N, Link W : Components and regulation of nuclear transport processes. FEBS J 2015; 282: 445–462.
Ibarra A, Hetzer MW : Nuclear pore proteins and the control of genome functions. Genes Dev 2015; 29: 337–349.
Simon DN, Rout MP : Cancer and the nuclear pore complex. Adv Exp Med Biol 2014; 773: 285–307.
Enarson P, Rattner JB, Ou Y, Miyachi K, Horigome T, Fritzler MJ : Autoantigens of the nuclear pore complex. J Mol Med 2004; 82: 423–433.
Duarte-Rey C, Bogdanos D, Yang CY et al: Primary biliary cirrhosis and the nuclear pore complex. Autoimmun Rev 2012; 11: 898–902.
Lima FA, Moreira-Filho CA, Ramos PL et al: Decreased AIRE expression and global thymic hypofunction in Down syndrome. J Immunol 2011; 187: 3422–3430.
Li J, Li X, Hou R et al: Psoriatic T cells reduce epidermal turnover time and affect cell proliferation contributed from differential gene expression. J Dermatol 2015; 42: 874–880.
Faria AM, Levay A, Wang Y et al: The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 2006; 24: 295–304.
Toda E, Terashima Y, Esaki K et al: Identification of a binding element for the cytoplasmic regulator FROUNT in the membrane-proximal C-terminal region of chemokine receptors CCR2 and CCR5. Biochem J 2014; 457: 313–322.
Rabe B, Vlachou A, Panté N, Helenius A, Kann M : Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci USA 2003; 100: 9849–9854.
Schmitz A, Schwarz A, Foss M et al: Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog 2010; 6: e1000741.
Copeland AM, Newcomb WW, Brown JC : Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol 2009; 83: 1660–1668.
Fay N, Panté N : Nuclear entry of DNA viruses. Front Microbiol 2015; 6: 467.
Park N, Katikaneni P, Skern T, Gustin KE : Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 2008; 82: 1647–1655.
Kuss SK, Mata MA, Zhang L, Fontoura BM : Nuclear imprisonment: viral strategies to arrest host mRNA nuclear export. Viruses 2013; 18: 1824–1849.
Rajani KR, Pettit Kneller EL, McKenzie MO, Horita DA, Chou JW, Lyles DS : Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog 2012; 8: e1002929.
Campbell EM, Hope TJ : HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 2015; 13: 471–483.
Marini B, Kertesz-Farkas A, Ali H et al: Nuclear architecture dictates HIV-1 integration site selection. Nature 2015; 521: 227–231.
Sarathi V, Shah NS : Triple-A syndrome. Adv Exp Med Biol 2010; 685: 1–8.
Storr HL, Kind B, Parfitt DA et al: Deficiency of ferritin heavy-chain nuclear import in triple a syndrome implies nuclear oxidative damage as the primary disease mechanism. Mol Endocrinol 2009; 23: 2086–2094.
Jühlen R, Idkowiak J, Taylor AE et al: Role of ALADIN in Human Adrenocortical Cells for Oxidative Stress Response and Steroidogenesis. PLoS One 2015; 10: e0124582.
Kaneb HM, Folkmann AW, Belzil VV et al: Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet 2015; 24: 1363–1373.
Neilson DE, Adams MD, Orr CM et al: Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am J Hum Genet 2009; 84: 44–51.
Kinoshita Y, Ito H, Hirano A et al: Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 2009; 68: 1184–1192.
Basel-Vanagaite L, Muncher L, Straussberg R et al: Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann Neurol 2006; 60: 214–222.
Alazami AM, Patel N, Shamseldin HE et al: Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 2015; 10: 148–161.
Miyake N, Tsukaguchi H, Koshimizu E et al: Biallelic mutations in nuclear pore complex subunit NUP107 cause early-childhood-onset steroid-resistant nephrotic syndrome. Am J Hum Genet 2015; 97: 555–566.
D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW : A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 2012; 22: 446–458.
Lupu F, Alves A, Anderson K, Doye V, Lacy E : Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 2008; 14: 831–842.
Niu X, Hong J, Zheng X et al: The nuclear pore complex function of Sec13 protein is required for cell survival during retinal development. J Biol Chem 2014; 289: 11971–11985.
Zheng X, Yang S, Han Y et al: A.Loss of zygotic NUP107 protein causes missing of pharyngeal skeleton and other tissue defects with impaired nuclear pore function in zebrafish embryos. J Biol Chem 2012; 287: 38254–38264.
Grill B, Chen L, Tulgren ED et al: RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans. J Neurosci 2012; 32: 2628–2636.
Zhang X, Chen S, Yoo S et al: Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 2008; 135: 1017–1027.
Schwartz M, Travesa A, Martell SW, Forbes DJ : Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin. Nucleus 2015; 6: 40–54.
Kehat I, Accornero F, Aronow BJ, Molkentin JD : Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol 2011; 193: 21–29.
Fakhro KA, Choi M, Ware SM et al: Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA 2011; 108: 2915–2920.
Xu L, Pan L, Li J et al: Nucleoporin 35 regulates cardiomyocyte pH homeostasis by controlling Na+-H+ exchanger-1 expression. J Mol Cell Biol 2015; 7: 476–485.
Mariño-Enríquez A, Wang WL, Roy A et al: Epithelioid inflammatory myofibroblastic sarcoma: an aggressive intra-abdominal variant of inflammatory myofibroblastic tumor with nuclear membrane or perinuclear ALK. Am J Surg Pathol 2011; 35: 135–144.
Takeoka K, Okumura A, Maesako Y, Akasaka T, Ohno H : Crizotinib resistance in acute myeloid leukemia with inv(2)(p23q13)/RAN binding protein 2 (RANBP2) anaplastic lymphoma kinase (ALK) fusion and monosomy 7. Cancer Genet 2015; 208: 85–90.
Roberts KG, Morin RD, Zhang J et al: Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 2012; 22: 153–166.
Zhou MH, Yang QM : NUP214 fusion genes in acute leukemia (Review). Oncol Lett 2014; 8: 959–962.
Fornerod M, Boer J, van Baal S et al: Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 1995; 10: 1739–1748.
Van Vlierberghe P, van Grotel M, Tchinda J et al: The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 2008; 111: 4668–4680.
Sandén C, Ageberg M, Petersson J, Lennartsson A, Gullberg U : Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC Cancer 2013; 13: 440.
Ageberg M, Drott K, Olofsson T, Gullberg U, Lindmark A : Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis. Genes Chromosomes Cancer 2008; 47: 276–287.
Takeda A, Yaseen NR : Nucleoporins and nucleocytoplasmic transport in hematologic malignancies. Semin Cancer Biol 2014; 27: 3–10.
Chatel G, Desai SH, Mattheyses AL, Powers MA, Fahrenkrog B : Domain topology of nucleoporin Nup98 within the nuclear pore complex. J Struct Biol 2012; 177: 81–89.
Gough SM, Slape CI, Aplan PD : NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 2011; 118: 6247–6257.
Light WH, Freaney J, Sood V et al: A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 2013; 11: e1001524.
Pascual-Garcia P, Jeong J, Capelson M : Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep 2014; 9: 433–442.
Crescenzi B, Nofrini V, Barba G et al: NUP98/11p15 translocations affect CD34+ cells in myeloid and T lymphoid leukemias. Leuk Res 2015; 39: 769–772.
Saw J, Curtis DJ, Hussey DJ, Dobrovic A, Aplan PD, Slape CI : The fusion partner specifies the oncogenic potential of NUP98 fusion proteins. Leuk Res 2013; 37: 1668–1673.
Greenblatt S, Li L, Slape C et al: Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood 2012; 119: 2883–2894.
Xu H, Menendez S, Schlegelberger B et al: Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13-driven mouse model. Blood 2012; 120: 3089–3097.
Petit A, Ragu C, Soler G et al: Functional analysis of the NUP98-CCDC28A fusion protein. Haematologica 2012; 97: 379–387.
Puthiyaveetil AG, Reilly CM, Pardee TS, Caudell DL : Non-homologous end joining mediated DNA repair is impaired in the NUP98-HOXD13 mouse model for myelodysplastic syndrome. Leuk Res 2013; 37: 112–116.
Funasaka T, Nakano H, Wu Y et al: RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle 2011; 10: 1456–1467.
Salsi V, Ferrari S, Gorello P et al: NUP98 fusion oncoproteins promote aneuploidy by attenuating the mitotic spindle checkpoint. Cancer Res 2014; 74: 1079–1090.
Singer S, Zhao R, Barsotti AM et al: Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell 2012; 48: 799–810.
Denk D, Bradtke J, König M, Strehl S : PAX5 fusion genes in t(7;9)(q11.2;p13) leukemia: a case report and review of the literature. Mol Cytogenet 2014; 7: 13.
Fortschegger K, Anderl S, Denk D, Strehl S : Functional heterogeneity of PAX5 chimeras reveals insight for leukemia development. Mol Cancer Res 2014; 12: 595–606.
Wang L, Motoi T, Khanin R et al: Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer 2012; 51: 127–139.
Hashizume C, Nakano H, Yoshida K, Wong RW : Characterization of the role of the tumor marker Nup88 in mitosis. Mol Cancer 2010; 9: 119.
Takahashi N, van Kilsdonk JW, Ostendorf B et al: Tumor marker nucleoporin 88 kDa regulates nucleocytoplasmic transport of NF-kappaB. Biochem Biophys Res Commun 2008; 374: 424–430.
Acknowledgements
We thank Dr Geraldine A Boyd for assistance with the manuscript and Dr Roberta Paolucci for Figure 1. This study was partly supported by Fondo per gli investimenti della Ricerca di Base FIRB 2011 RBAP11TF7Z_005; Associazione Italiana per la Ricerca sul Cancro AIRC IG-15525 and Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale PRIN 2010NYKNS7_003.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Nofrini, V., Di Giacomo, D. & Mecucci, C. Nucleoporin genes in human diseases. Eur J Hum Genet 24, 1388–1395 (2016). https://doi.org/10.1038/ejhg.2016.25
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.25
This article is cited by
-
Downregulation of NUP93 aggravates hypoxia-induced death of cardiomyocytes in vitro through abnormal regulation of gene transcription
Acta Pharmacologica Sinica (2023)
-
Early-life exercise primes the murine neural epigenome to facilitate gene expression and hippocampal memory consolidation
Communications Biology (2023)
-
Architecture and composition of plant nucleopore complexes, comparisons with putative homologs across kingdoms
The Nucleus (2023)
-
Elitist random swapped particle swarm optimization embedded with variable k-nearest neighbour classification: a new PSO variant applied to gene identification
Soft Computing (2023)
-
Genes of the Ubiquitin Proteasome System Qualify as Differential Markers in Malignant Glioma of Astrocytic and Oligodendroglial Origin
Cellular and Molecular Neurobiology (2023)