Abstract
The promise of personalized medicine depends on the ability to integrate genetic sequencing information into disease risk assessment for individuals. As genomic sequencing technology enters the realm of clinical care, its scale necessitates answers to key social and behavioral research questions about the complexities of understanding, communicating, and ultimately using sequence information to improve health. Our study captured the motivations and expectations of research participants who consented to participate in a research protocol, ClinSeq, which offers to return a subset of the data generated through high-throughput sequencing. We present findings from an exploratory study of 322 participants, most of whom identified themselves as white, non-Hispanic, and coming from higher socio-economic groups. Participants aged 45–65 years answered open-ended questions about the reasons they consented to ClinSeq and about what they anticipated would come of genomic sequencing. Two main reasons for participating were as follows: a conviction to altruism in promoting research, and a desire to learn more about genetic factors that contribute to one's own health risk. Overall, participants expected genomic research to help improve understanding of disease causes and treatments. Our findings offer a first glimpse into the motivations and expectations of individuals seeking their own genomic information, and provide initial insights into the value these early adopters of technology place on information generated by high-throughput sequencing studies.
Similar content being viewed by others
Introduction
Genomics has already revolutionized the biological sciences, and the future application of genomics to health care has the potential to improve prevention, diagnosis, and treatment of disease by refining individual risk in the clinical setting. However, this potential can only be realized through translational and clinical research studies to establish the relationship of disease risk to genomic variants derived from sequencing data. These studies will necessitate the enrollment of thousands of clinical research participants, and thus methods and approaches need to be developed to interact and communicate with participants regarding clinical genomics. As well, these clinical genomics studies will serve as the foundation for the development of approaches to health-care providers’ interactions with patients undergoing sequencing in the clinic and hospital wards of the future.
High-throughput genomic sequencing can elucidate an enormous range of sequence and copy number variations for a given individual. A typical whole-genome sequence determination yields on the order of 4 000 000 sequence variations that differ from the current human reference sequence. While most are benign or of unknown consequence, some are associated with a significant increased risk of disease for the individual and/or their family members. It is important to distinguish sequencing from common variant detection (eg, SNP chips). Sequencing can detect rare variants that are associated with relative risks that approach the inverse of the disease frequency (ie, as high as 500 for a disease such as familial hypercholesterolemia), whereas SNP typing typically detects variants that yield relative risks of 1.05–1.2. The typical human is a carrier for three to five deleterious genetic variants or mutations that cause severe recessive diseases.1, 2 In addition, there are over 30 known cancer susceptibility syndromes, which in aggregate may affect more than 1/500 people, and the sequence variants that cause these disorders can be readily detected with whole-genome sequencing. Rare variants in the causative genes can predict very high relative risks, and thus have significant potential medical impact and clinical utility. Other heritable conditions that affect more than 1/500 people of Caucasian extraction include the hypertrophic cardiomyopathies3 and hemochromatosis.4 All patients/research participants are likely to have several deleterious variants, and it will be important for clinicians to be familiar with the motivations and expectations of patients regarding the return of such results.
The translation of genomics into useful and cost-effective clinical care will require years of translational research. The early phases of translational research, often referred to as proof-of-principle studies, are typically characterized by the introduction of new technologies to normal volunteers and carefully defined populations. Anticipating the early phase of translational genomics research, a recent analog study assessed the views of 4659 participants from the general public.5 They were asked about their attitudes toward participating in a theoretical genetic–environment research biobank, specifically their opinions about controlling access to their theoretical samples and the information that could be derived from them. The participants expressed a clear desire for ongoing choices and control regarding the use of their samples and the information that is derived from their samples. This important study is nevertheless limited by its analog design. An important next step is to replicate the findings in actual research participants. Here we extend the results of this analog study by exploring the motivations and expectations of participants who consented to donate their DNA to an actual clinical genomic research study.
The context in which this exploratory study was performed is a clinical genomic protocol, known as the ClinSeq study.6 The ClinSeq study aims to enroll a cohort of more than 1000 participants who consent to full-genome sequencing. The participants are broadly consented, up to and including whole-genome sequencing and future exploration of any and all possible phenotypes. It is, therefore, an ideal cohort for studying motivations and expectations because the possibilities for receiving results are so wide ranging. This study provides initial findings regarding the motivations and expectations of individuals choosing to have their whole genome or exome sequenced, with the option of learning about individual genotype results in the future.
Methods
Survey data were collected from 322 individuals enrolled in the ClinSeq study between January 2007 and May 2008. Individuals eligible for the ClinSeq study were between 45 and 65 years of age, and most were local to the Washington, DC and Baltimore, Maryland metropolitan areas. Participants were not offered financial compensation. The target ClinSeq cohort of 1000 individuals will include participants with a range of risk to develop coronary artery disease (CAD), including asymptomatic individuals with varying degrees of risk to develop CAD and participants with known CAD. The group with known CAD includes individuals with a previous history of myocardial infarction, silent myocardial infarction, stent placement, revascularization, or 50% or more arterial blockage. Out of the 322 participants who completed the survey, 294 (91.3%) were asymptomatic for CAD, and 28 (8.7%) had a history of CAD at the time of enrollment. From its inception the goal of ClinSeq has been to sequence most or all regions of 1000 human genomes, and more than 200 exome sequences had been completed at the time of publication. Details on the ClinSeq cohort are described elsewhere.2 The National Human Genome Research Institute Institutional Review Board approved this study.
During an enrollment visit and before consenting to whole-genome sequencing and participating in genetic counseling, participants completed a baseline survey. Reasons/motivations for participating in ClinSeq, expectations of whole-genome sequencing and socio-demographic characteristics were assessed.
Motivations
To assess motivations, respondents were asked, ‘What are your reasons for wanting to participate in this study’?
Expectations
A multiple-choice question was used to assess participants’ expectations of whole-genome sequencing.
Testing for many genes can… (Check all that apply)
-
Find a genetic risk for a disease that you do not have, but could develop in the future
-
Find a genetic cause or contribution for a disease that you have
-
Give you a ‘clean bill of health’
-
Give you information about not only you, but also your relatives
-
None of the above
-
Don’t know
To further assess expectations, they were asked, ‘What else, if anything, could be learned from testing many genes’?
Responses to the open-ended questions were read for content and an initial codebook was developed. A systematic review of the literature on ‘diffusion of technology, early adopters of technology’, ‘individuals’ motivations to undergo genetic testing’, and ‘individuals’ reasons to become involved with research in general’ provided empirical data for the concepts included in the final codebook. It was revised for content validity through an iterative process involving discussions among the research team until the coding scheme was determined to be concise and robust. Using NVIVO 7 QSR, a qualitative analysis software package, the responses were analyzed by two independent coders to ensure coding consistency. One member of the research team coded the responses to the motivation question and approximately 25% were randomly selected and coded by a second coder. Similarly, one primary coder coded responses to the expectation question and a second coder coded approximately 33% of the responses. Inter-coder reliability was determined to be 95% for the motivation question and 96% for the expectation question. Discrepancies that emerged between the coders were discussed and reconciliation was achieved. Data saturation was facilitated by the significant sample size.
We identified common themes among answers to both questions. These themes were correlated with health-related attributes to determine whether there were relationships between the two – for example, whether people with symptoms of CAD were more likely than healthy volunteers to participate because of a hope for personal health benefits. There were no differences between the two groups of ClinSeq participants. Responses to the multiple-choice question were summed. Similarly, there were no differences between the two groups of participants.
Results
Study population
Between January 2007 and May 2008, a total of 337 participants enrolled in the ClinSeq study. Out of the 337 enrollees, 322 (96%) completed the survey. Table 1 summarizes the participants’ characteristics. The majority of the 322 participants were white, not of Hispanic or Latino background, and from higher socio-economic groups. The average age was 57 years and the female to male ratio approached one.
Motivations to enroll in a whole-genome sequencing study
Of the 322 participants, 313 (97%) provided responses about their reasons for participating in the study. Two main themes were identified in the responses: a conviction to altruism in promoting research, and a desire to learn more about factors that contribute to their own health. These themes originated from separate respondents with minimal overlap. The two groups were similar with regard to their socio-demographic characteristics and their CAD status.
Altruism
Out of the 313 participants, 141 (44%) responded in an altruistic manner. Although the majority of these responses were general in nature – ‘Help someone who may be at risk for CAD or other related disorders’ or ‘Help others in the future’, some addressed an intent to contribute to research about genetics (26/313) or research about health (33/313). For example, participants mentioned that they were enrolling in the study ‘For the contribution to a major effort to correlate disease/risk and gene sequence on the global level’ and ‘Because I can give back and be part of groundbreaking (and potentially life saving) research’.
Seeking personal health information
Fifty-six percent (175/313) mentioned that they were seeking personal health information. A proportion of these responses (63/313) reflected participants’ desire to learn information about a specific health condition, as shown in the following response: ‘I became familiar with isolating the gene specific to Dystonia, which my son has. When my colleague at work mentioned this study, I was interested’. Out of this subgroup, the majority was seeking information about heart disease – ‘To learn about my health and risk of heart disease. There is a family history of hypertension and heart disease’. Some participants were interested in receiving information about genetic risk (41/313) and predisposition to disease (20/313), as illustrated by the following quotes: ‘To learn about my health and risk for not only CAD, but other disease processes as well’ and ‘Interesting study. Would like to know if I have genes that predispose to disease’. Other participants had concerns related to their family history of disease (38/313) – ‘Curious about genetic predisposition in family, especially as it relates to family history of autoimmune disease’.
Future uses of genomic information
Other themes emerged from the responses to the motivation question. Thirteen percent (42/313) of the responses reflected some of the participants’ desire to learn information that may be helpful to their relatives. This theme of seeking information for family is illustrated in responses like – ‘Family cardiac history and knowledge that would help me make better choices for my health and my children’ and ‘I want to learn if my children and other posterity may be at risk for heart disease, diabetes, and other diseases’.
Some participants also described future benefits of participating in the study (50/313), including specifics such as the potential for preventing disease (14/313) – ‘Provide me with information to prevent development or lessen the impact of some medical conditions’. Additionally, a number cited the potential for finding better treatment (11/313), hinting at the concept of pharmacogenomics. Within this notion of future benefits, a small number of participants mentioned finding cures for diseases – ‘I believe that genetics is the place in science that offers the most promising future for disease prediction and cure’.
Finally, some participants mentioned curiosity (59/313) as a reason for participation, as illustrated by the following responses – ‘To be on the cutting edge of this information’ and ‘Curiosity, continuous learning. I work in health care field and am fascinated by research capability’.
Expectations from a whole-genome sequencing study
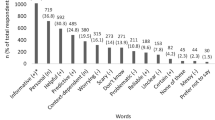
Out of 322 participants who completed the survey, 191 (59%) provided responses about their expectations of whole-genome sequencing. Most responses (142/191) reflected a sense that genomic information could lead to a better understanding of disease causes and treatments. Specifically, 74/191 had expectations that whole-genome sequencing would increase knowledge of the causes and development of disease – ‘Why some people develop diseases that do not have a genetic predisposition or why some who do have a genetic predisposition do not develop the disease’. Fifty percent of these respondents expected to gain knowledge of the relationship between genes and disease, such as the identification of predispositions or risk factors for disease – ‘To determine if diseases may develop in different ways according to gene differences’.
Some individuals felt that genomic information could lead to the advancement of treatment, diagnoses, or cures, and more specifically to the advancement of tailored or personalized medicine (53/191), including pharmacogenomics (12/191). The following illustrate these expectations – ‘Designing personal drug therapies’ and ‘Possible link to a medicine to treat a disease’.
Reflecting scientific literacy, about 23% (33/191) of the respondents described the contribution of whole-genome sequencing research to the understanding of genetic mechanisms, such as the role of gene–gene interactions. They described learning the ‘Relationships of particular genes to diseases and the interactions of genes to cause or inhibit disease’, and ‘Interactions that provide protective factors or benefits (such as sickle cell anemia versus malaria resistance)’.
Multiple-choice question regarding expectations
The response rate was 100% (322/322) for the multiple-choice question in the survey. Sixty-eight percent (218/322) of the participants answered the question with reasonable expectations, checking the three following answers regarding what could be learned from testing many genes: find a genetic risk for a disease that you do not have, but could develop in the future, find a genetic cause or contribution for a disease that you have, and give you information about not only you, but also your relatives. Of note, 26 out of 322 (8%) participants also chose the option give you a ‘clean bill of health’ as a response.
Discussion
High-throughput genomic research is characterized by a number of unique attributes, including the enormous scale of the data sets generated for a single individual, and the wide range of possible genetic results. Additionally, genomic research introduces a new paradigm in the search and delivery of genetic information, going directly from the genotype to the phenotype. These novel characteristics dictate a need to discern the motivations and expectations of research participants choosing to have their whole genome or exome sequenced.
The results of this exploratory study reveal that altruism is one of the major sources of motivation for ClinSeq participants. The motivation of altruism echoes that of other healthy research volunteers reported in the literature.7 Although financial compensation is the greatest motivation for volunteering in phase I or proof-of-principle studies, it is less valued as a motivator by healthy volunteers with higher income and education8 and older age.7 Altruism and personal health gain/benefit have been reported as particularly important motivators for volunteers of older ages.7 Among genetic studies that do not disclose individual results, there are parallel findings. Healthy volunteers have reported financial compensation as an important motivator; however, age has been inversely related to the importance of financial compensation on the decision to participate.9 Individuals affected with a specific genetic condition have reported participating in genetic studies for altruistic reasons: to help to find a gene, to help others who have or are at-risk for disease, and to help raise awareness of disease.10
In contrast, personal health gain has been reported as the greatest motivation for patients to enroll in clinical trials of cancer chemotherapy.11, 12, 13 Cancer patients may anticipate that they will derive direct benefit, even when they understand that a study is unlikely to impart any personal benefit. This is most often seen when participants’ prognoses are terminal or the technology under study is viewed as ‘cutting edge’. This motivation has been described as the ‘therapeutic misconception’14, 15 and depicts the hope that research participants may experience when clinical treatments have failed to help them. In this study, the participants were healthy volunteers and patients with symptoms of coronary heart disease. Their overall health status was not as dire as that of cancer patients participating in phase-I clinical trials, thus personal health gain for ClinSeq participants is likely to carry a different meaning. In fact, the participants share more in common with early adopters of new technologies and with individuals who undergo genetic testing, than with clinical research participants who are gravely ill.
Early adaptation of new technologies typically proceeds in the face of considerable uncertainty about the potential indications, risks, and usefulness.16 Innovators and early adopters are more often information seekers and risk takers with higher formal education and higher financial status.17 They show tolerance of the uncertainties. This sophistication is reflected among the participants in this study in their expectations of whole-genome sequencing. They articulated how results may improve understanding of the cause of disease and lead to improved treatments, expectations that are consistent with the potential information that individuals could learn from having their genome sequenced.18 This understanding was further evidenced by their reasons for participating, with many hoping to learn about their risks for certain diseases, and some expecting to learn about genetic predispositions that they could possibly pass on to future offspring.
Early adopters of clinical genetic tests share characteristics with ClinSeq participants. A retrospective study of women offered BRCA1/2 testing soon after it became available, and found that uptake was associated with higher innovativeness and greater congruence between the test and their existing values. These findings are compatible with hypotheses generated by the diffusion of innovation theory.19
Similar to other studies,19, 20 most ‘early adopters’ in this study were more likely to have a college degree or higher. ClinSeq participants share motivations with individuals who enroll in research in general, as well as with those who typically come forth for genetic studies and new genetic tests based on their family or personal history of disease. Individuals afflicted with a hereditary condition undergo genetic testing to provide information to family members,21, 22 gain information for self in terms of disease management,22, 23, 24 to reduce uncertainty,23 and for altruistic reasons.22 Those undergoing genetic testing because they are at risk for a specific hereditary condition cite equivalent motivations.25, 26, 27, 28, 29, 30 Interestingly, these individuals feel that in addition to providing them with the ability to guide future health-care decisions, genetic test results also aid in planning for future non-medical decisions, such as preparing advance directives and enrolling in long-term insurance.25, 26, 27, 28, 29
Overall, the findings from this study suggest that a significant proportion of early participants in whole-genome sequencing studies expect to gain information about their health risks. Taken together with the findings from an analog study,5 this suggests that participants also value having a choice about what information they can learn. When designing whole-genome sequencing studies, successful recruitment may depend on offering a choice to participants about the return of results. To do so, studies will need to anticipate providing access to clinical research support resources.31
The results of this study are limited by the characteristics of the ClinSeq cohort, which is comprised largely of individuals of higher levels of formal education and high socio-economic status. Although our findings parallel those of other early adopters, they cannot be generalized to other socio-demographic populations. The use of a multiple-choice question about participants’ expectations preceding the open-ended question may have biased responses. Yet we found participants’ responses replicate findings of other genetic testing and research participation studies.5
In summary, sequencing technology is predicted to continue advancing at a very rapid pace, along with further reductions in cost. In the not too distant future, whole-genome sequencing will enter clinical care. ClinSeq provides a timely opportunity to explore the motivations and expectations of participants anticipating return of high-throughput, whole-genome sequencing results in the context of translational research. Additional ClinSeq studies are ongoing to measure participants’ attitudes toward intentions and decisions to learn results.
References
Morton NE : The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am J Hum Genet 1956; 8: 80–96.
Morton NE : The mutational load due to detrimental genes in man. Am J Hum Genet 1960; 12: 348–364.
Charron P, Arad M, Arbustini E et al: Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2010; 31: 2715–2726.
Adams PC, Reboussin DM, Barton JC et al: Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med 2005; 352: 1769–1778.
Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K : Public perspectives on informed consent for biobanking. Am J Public Health 2009; 99: 2128–2134.
Biesecker LG, Mullikin JC, Facio FM et al: The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res 2009; 19: 1665–1674.
van Gelderen CE, Savelkoul TJ, van Dokkum W, Meulenbelt J : Motives and perception of healthy volunteers who participate in experiments. Eur J Clin Pharmacol 1993; 45: 15–21.
Almeida L, Azevedo B, Nunes T, Vaz-da-Silva M, Soares-da-Silva P : Why healthy subjects volunteer for phase I studies and how they perceive their participation? Eur J Clin Pharmacol 2007; 63: 1085–1094.
McCarty CA, Nair A, Austin DM, Giampietro PF : Informed consent and subject motivation to participate in a large, population-based genomics study: the Marshfield Clinic Personalized Medicine Research Project. Community Genet 2007; 10: 2–9.
Treloar SA, Morley KI, Taylor SD, Hall WD : Why do they do it? A pilot study towards understanding participant motivation and experience in a large genetic epidemiological study of endometriosis. Community Genet 2007; 10: 61–71.
Nurgat ZA, Craig W, Campbell NC, Bissett JD, Cassidy J, Nicolson MC : Patient motivations surrounding participation in phase I and phase II clinical trials of cancer chemotherapy. Br J Cancer 2005; 92: 1001–1005.
Schutta KM, Burnett CB : Factors that influence a patient's decision to participate in a phase I cancer clinical trial. Oncol Nurs Forum 2000; 27: 1435–1438.
Moore S : A need to try everything: patient participation in phase I trials. J Adv Nurs 2001; 33: 738–747.
Henderson GE, Easter MM, Zimmer C et al: Therapeutic misconception in early phase gene transfer trials. Soc Sci Med 2006; 62: 239–253.
Sulmasy DP, Astrow AB, He MK et al: The culture of faith and hope: patients’ justifications for their high estimations of expected therapeutic benefit when enrolling in early phase oncology trials. Cancer 2010; 116: 3702–3711.
Gelijns A : Lessons for genomics from other technologies; in: Diffusion and use of Genomic Innovations in Health and Medicine: Workshop Summary. Washington, DC: National Academies Press, 2008, p 17.
Rogers EM : Diffusion of Innovations, 5th eds New York: Free Press, 2003.
Ashley EA, Butte AJ, Wheeler MT et al: Clinical assessment incorporating a personal genome. Lancet 2010; 375: 1525–1535.
Armstrong K, Weiner J, Weber B, Asch DA : Early adoption of BRCA1/2 testing: who and why. Genet Med 2003; 5: 92–98.
Hobbs P, Smith A, George WD, Sellwood RA : Acceptors and rejectors of an invitation to undergo breast screening compared with those who referred themselves. J Epidemiol Community Health 1980; 34: 19–22.
Manne SL, Chung DC, Weinberg DS et al: Knowledge and attitudes about microsatellite instability testing among high-risk individuals diagnosed with colorectal cancer. Cancer Epidemiol Biomarkers Prev 2007; 16: 2110–2117.
Esplen MJ, Madlensky L, Aronson M et al: Colorectal cancer survivors undergoing genetic testing for hereditary non-polyposis colorectal cancer: motivational factors and psychosocial functioning. Clin Genet 2007; 72: 394–401.
Sankar P, Wolpe PR, Jones NL, Cho M : How do women decide? Accepting or declining BRCA1/2 testing in a nationwide clinical sample in the United States. Community Genet 2006; 9: 78–86.
Balmana J, Stoffel EM, Emmons KM, Garber JE, Syngal S : Comparison of motivations and concerns for genetic testing in hereditary colorectal and breast cancer syndromes. J Med Genet 2004; 41: e44.
Gooding HC, Linnenbringer EL, Burack J, Roberts JS, Green RC, Biesecker BB : Genetic susceptibility testing for Alzheimer disease: motivation to obtain information and control as precursors to coping with increased risk. Patient Educ Couns 2006; 64: 259–267.
Claes E, Denayer L, Evers-Kiebooms G, Boogaerts A, Legius E : Predictive testing for hereditary non-polyposis colorectal cancer: motivation, illness representations and short-term psychological impact. Patient Educ Couns 2004; 55: 265–274.
Hurley AC, Harvey FR, Roberts JS et al: Genetic susceptibility for Alzheimer's disease: why did adult offspring seek testing? Am J Alzheimers Dis Other Demen 2005; 20: 374–381.
Esplen MJ, Madlensky L, Butler K et al: Motivations and psychosocial impact of genetic testing for HNPCC. Am J Med Genet 2001; 103: 9–15.
Roberts JS, LaRusse SA, Katzen H et al: Reasons for seeking genetic susceptibility testing among first-degree relatives of people with Alzheimer disease. Alzheimer Dis Assoc Disord 2003; 17: 86–93.
Fraser L, Bramald S, Chapman C et al: What motivates interest in attending a familial cancer genetics clinic? Fam Cancer 2003; 2: 159–168.
Ormond KE, Wheeler MT, Hudgins L et al: Challenges in the clinical application of whole-genome sequencing. Lancet 2010; 375: 1749–1751.
Acknowledgements
This study was supported by funding from the Intramural Research Program of the National Human Genome Research Institute. We thank Riley Cooper-McCann and Haley Eidem for providing assistance in compiling the data. Most importantly, we thank the study participants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Facio, F., Brooks, S., Loewenstein, J. et al. Motivators for participation in a whole-genome sequencing study: implications for translational genomics research. Eur J Hum Genet 19, 1213–1217 (2011). https://doi.org/10.1038/ejhg.2011.123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.123
Keywords
This article is cited by
-
Evaluation of CTRL: a web application for dynamic consent and engagement with individuals involved in a cardiovascular genetic disorders cohort
European Journal of Human Genetics (2024)
-
Lessons learned during the process of reporting individual genomic results to participants of a population-based biobank
European Journal of Human Genetics (2023)
-
Parents, their children, whole exome sequencing and unsolicited findings: growing towards the child’s future autonomy
European Journal of Human Genetics (2021)
-
Patients’ decision to contribute to a biobank in the light of the patient-recruiter relationship—a qualitative study of broad consent in a hospital setting
Journal of Community Genetics (2021)
-
Just data? Solidarity and justice in data-driven medicine
Life Sciences, Society and Policy (2020)