Abstract
Mitochondrial complex I deficiency is the most common defect of the oxidative phosphorylation system. We report a patient with Leigh syndrome who showed a complex I deficiency expressed in cultured fibroblasts and muscle tissue. To find the genetic cause of the complex I deficiency, we screened the mitochondrial DNA and the nuclear-encoded subunits of complex I. We identified compound-heterozygous mutations in the NDUFA10 gene, encoding an accessory subunit of complex I. The first mutation disrupted the start codon and the second mutation resulted in an amino acid substitution. The fibroblasts of the patient displayed decreased amount and activity, and a disturbed assembly of complex I. These results indicate that NDUFA10 is a novel candidate gene to screen for disease-causing mutations in patients with complex I deficiency.
Similar content being viewed by others
Introduction
NADH:ubiquinone oxidoreductase (E.C.1.6.5.3.), or complex I is the first and largest of the five complexes of the oxidative phosphorylation (OXPHOS) system. Its function is binding and oxidizing NADH to free electrons, which are then transferred to the electron acceptor ubiquinone. The energy released during this electron transfer is used to translocate protons across the inner mitochondrial membrane, generating a proton gradient, which can be used for the synthesis of ATP. Complex I consist of 45 subunits out of which 7 are encoded by the mitochondrial DNA (mtDNA). It is an L-shaped complex, consisting of a hydrophobic membrane arm embedded in the mitochondrial inner membrane and a hydrophilic peripheral arm protruding into the matrix. The complex can be divided into three functional modules. The dehydrogenase module is important for the oxidation of NADH, the hydrogenase module has a role in the transport of electrons to ubiquinone, and the proton translocation module is involved in proton pumping.1, 2
Isolated complex I deficiency is the most common defect of the OXPHOS system, accounting for approximately 23% of all patients with childhood respiratory chain deficiency.3 It has a wide clinical variety, affecting one or more tissues or organs.4 The organs with the highest energy demand such as heart, brain, skeletal muscle tissue, and liver are the most affected organs. Owing to the bi-genomic control of the OXPHOS system, mutations causing complex I deficiency can be found in either the mtDNA or in genes encoded by the nuclear DNA. Previous studies identified disease-causing mutations in nuclear structural genes encoding for the seven core subunits (NDUFS1, NDUFS2, NDUFS3, NDUFS7, NDUFS8, NDUFV1, and NDUFV2) and five accessory subunits of complex I (NDUFS4, NDUFS6, NDUFA1, NDUFA2, and NDUFA11).5, 6, 7, 8 Furthermore, mutations have been described in eight assembly factors (NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, C8orf38, C20orf7, ACAD9, and NDUBPL) of this complex and in an uncharacterized protein (FOXRED1) causing complex I deficiency.9, 10, 11, 12, 13, 14, 15, 16
Although pathogenic mutations have been described in accessory subunits, the function of these subunits is not exactly known yet. It has been suggested that some are important for the biogenesis of complex I. One of these subunits is NDUFA10. The predicted 355 amino acid human protein is 80% identical to the 42-kDa bovine homolog. This subunit is located in the hydrophobic protein fraction of complex I, and might therefore be involved in the transfer of protons. Furthermore, NDUFA10 is one of the subunits that undergoes post-translational modification; it can be phosphorylated at a single amino acid that is, serine 59 (Schulenberg et al17 and Schilling et al18).
In the present communication, we describe two compound heterozygous mutations in a patient with a complex I deficiency, expressed in cultured fibroblasts and muscle tissue. Cell biological studies were performed to show the functional consequences of the genetic variations. A new gene responsible for human complex I deficiency has been identified.
Subjects and methods
Case report
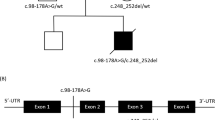
The patient, a boy, was born after a normal pregnancy of 32 weeks with a sectio caesaria because of fetal distress. His birth weight was 2715 g. He had a normal start and neonatal period. From early on, he showed hypotonia. His milestones were uneventful with regard to laughing, contact, grabbing things, and rolling over to his back, but he did not reach sitting position, and head control remained poor. At 10 months of age, he was referred for evaluation of the cause of his retarded development and hypotonia. Tendon reflexes were somewhat increased. Therefore, it was concluded that there was a central cause of hypotonia together with retarded development. His blood and cerebrospinal fluid lactate were 8.6 and 4.9 mmol/l, respectively (reference value 0.5–2.2 mmol/l), with increased lactate to pyruvate ratios (being around 20 on more than one occasion and the one measurement in cerebrospinal fluid). His cerebral MRI showed symmetrical lesions in especially the basal ganglia and substantia nigra. On the basis of the high lactate concentrations and the increased lactate/pyruvate ratio, a defect of pyruvate dehydrogenase complex or within the OXPHOS was likely considered. Biochemical investigations were performed in muscle and fibroblasts (Table 1). We started with thiamine and a ketogenic diet administered by gastrointestinal tube feeding. Owing to analyses of blood gases, showing a pH of 7.12 with 4 mmol/l of bicarbonate, sodium bicarbonate was given, resulting in normalization of pH with some respiratory compensation (pCO2 usually between 3.5 and 4.0 kPa). With these strategies, growth showed adequate response, so that his early fragility disappeared. His development improved with uttering noises rather than clear words. During later months, he displayed periods of abnormal breathing with hypo- and hyperventilation. At one of the occasions, he seemed to have a convulsion. Routine cardiac evaluation by ultrasound suggested some hypertrophic cardiomyopathy, without clear clinical relevance at that moment. During one such episode of convulsions, he experienced a severe cardiac respiratory arrest. X-ray of the thorax showed massive pleural fluid, most probably as a result of cardiac decompensation by hypertrophic cardiomyopathy. Notwithstanding treatment with mechanical ventilation, with high pressures, and the use of cardiovascular agents to stimulate contractility followed by resuscitation procedures, were unsuccessful; he died at 23 months of age. In his last days, he also showed hypoglycaemia (glucose 1.6 mmol/l), a finding not being observed before, possibly because of multiorgan failure rather than isolated liver failure. At autopsy, besides the already known cardiomyopathy, degenerative abnormalities were reported in the liver, spleen, kidneys, and pancreas. Apart from these findings, the most outspoken abnormalities were seen in the brain: in the thalamus, the pons, and also in the hippocampus degradation of myeline, decreased myelinization, proliferation of vessels, some gliosis, and some foam cell response were also reported. Especially from the abnormalities found in the thalamus and the pons, the diagnosis of classical Leigh syndrome was concluded.
Cell culture and biochemical analysis
Skin fibroblasts were cultured in M199 medium (Life Technologies, Breda, The Netherlands) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin in a humified atmosphere of 95% air and 5% CO2 at 37°C. Measurement of the mitochondrial respiratory chain enzyme activities in muscle tissue and skin fibroblasts were performed according to established procedures.19, 20 The values were expressed relative to the activity of cytochrome c oxidase and/or citrate synthase.
Genetic analysis of the mtDNA and nuclear complex I genes
Patient DNA was extracted from cultured skin fibroblasts by standard procedures.21 Oligonucleotide primers were designed from the mtDNA sequence and from the gene sequences of all nuclear-encoded complex I subunits. The Genbank accession number of the NDUFA10 gene sequence is NM_004544.3. Other Genbank accession numbers and primer sequences are available on request. The size of the amplification products were checked with a 1.5% agarose gel and the amplimers were sequenced directly with the BigDye terminator cycle sequencing chemistry on an Applied Biosystems (Nieuwerkerk a/d IJssel, The Netherlands) ABI PRISM 3130xl Genetic Analyzer. The nucleotide numbering follows cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1.
BN-PAGE analysis and in-gel activity assay
Blue native-gradient gels (5–15%) were cast and run with 40 μg of digitonin-isolated mitochondria as described previously.22, 23 After electrophoresis, the gels were further processed for in-gel activity assay or two-dimensional 10% SDS-PAGE, followed by immunoblot analysis as described by Calvaruso et al.22
SDS-PAGE analysis
Mitochondrial lysates were separated on a 10% SDS-PAGE gel. Next, the proteins were transferred to PROTAN nitrocellulose membrane (Schleicher & Schuell, Zwijndecht, The Netherlands) for immunodetection.
Antibodies and ECL detection
Western blotting was performed with primary antibodies raised against the complex I subunits NDUFA9 (Invitrogen, Breda, The Netherlands), NDUFS3 (Invitrogen), NDUFA10 (Sigma-Aldrich, Zwijndrecht, The Netherlands), ND1 (Dr A Lombes, Paris), and the complex II 70 kDa subunit (SDHA; Invitrogen). Secondary antibodies used were peroxidase-conjugated anti-mouse (Invitrogen), anti-rabbit (Invitrogen), or anti-chicken IgGs (Southern Biotech, Birmingham, AL, USA), and detection of the signal was performed using ECL Plus (Amersham Biosciences, Roosendaal, The Netherlands) according to the manufacturer's instructions.
Results
Biochemical studies
Investigation of the activities of the respiratory enzyme complexes in cultured fibroblasts and fresh muscle tissue of the index patient revealed a severely reduced complex I activity (Table 1). The enzymatic activities were 29 and 7% from the lowest value of the control range in the fibroblasts and muscle tissue, respectively. Furthermore, the activity of complex III in muscle tissue was slightly decreased (68% of the lowest control value).
Molecular genetic studies in the NDUFA10 gene
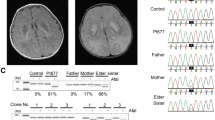
We searched for the molecular defect causing the complex I deficiency by screening the entire mtDNA, followed by screening the genes for all 38 nuclear-encoded subunits. We excluded the presence of disease-causing mutations in the mtDNA. Sequence analysis of the nuclear NDUFA10 gene demonstrated the presence of two heterozygous substitutions (Figure 1a). The first mutation is a substitution from A-to-G (c.1A>G) in exon 1, which disrupts the start codon methionine (p.Met1?). The next possible start codons can be found at 116 and 118 nucleotides upstream. However, these alternative start codons are either not in frame or abolish the mitochondrial target sequence, which makes both unlikely to be functional. The father of the index patient was carrier of this mutation. The second mutation was found in exon 3 (c.425A>G), causing a substitution of amino acids (p.Gln142Arg). This mutation affects a highly conserved amino acid (Figure 1b), and the mutation software SIFT (J. Craig Venter Institute, Rockville, MD, USA) predicted this mutation to be pathogenic because it affects the protein function with a score of 0.00. Substitutions with score less than 0.05 are predicted to affect protein function. The mother of the patient was found to be heterozygous for this mutation. Both the mutations were not present in 108 healthy controls or 21 other patients with isolated complex I deficiency expressed in cultured fibroblasts. From these observations, we conclude that it is highly likely that both mutations are pathogenic.
Molecular analysis of the NDUFA10 gene. (a) Electropherograms showing the wild-type sequence of NDUFA10 (top) and the nucleotide changes in fibroblasts of the patient (bottom). Arrows represent the nucleotide substitutions, c.1A>G (left) and c.425A>G (right). (b) Conservation of the altered amino acid (p.Gln142Arg) is shown in a Clustal W alignment. Abbreviations : Wt, wild-type; P, index patient.
Effect of the NDUFA10 mutations on the activity and steady-state level of complex I
The effect of the mutations on the activity of complex I was examined by performing BN-PAGE analysis followed by western blotting with an anti-NDUFA9 antibody to detect complex I amounts, and in-gel complex I activity measurements with mitochondria-enriched fractions of the cultured patient fibroblasts. Figure 2a shows a marked decrease of complex I activity in patient fibroblasts compared with the control cells. Furthermore, the expression of NDUFA9, indicative of the amount of fully-assembled complex I, was severely decreased. These results are in agreement with the spectrophotometric complex I observations in the patient fibroblasts.
The mutations in NDUFA10 result in decreased activity and amount of complex I. (a) BN-PAGE analysis followed by an in-gel activity assay demonstrates a decreased complex I activity in fibroblasts of the patient compared with control cells. The amount of subunit NDUFA9 is also severely reduced, indicating a decreased amount of whole complex I. The amount of complex II is not affected. (b) SDS-PAGE analysis and immunoblotting with an antibody directed against NDUFA10 shows that the amount of NDUFA10 is decreased in patient fibroblasts. The amount of NDUFA9 is also reduced, suggesting a reduction in amount of complex I. The amount of complex II is not affected. Abbreviations: C, control; P-A10, patient with NDUFA10 mutations; C, I, II, complexes I, II; IGA, in-gel activity; SDHA, succinate dehydrogenase complex subunit A.
Next, we performed SDS-PAGE analysis to study the effect of the mutations on the expression of complex I. Figure 2b shows a clear decrease in the protein level of NDUFA10 in the patient compared with the control. The expression of the NDUFA9 subunit was also decreased.
Effect of the NDUFA10 mutations on the assembly of complex I
To test whether the assembly of complex I is disturbed, we performed two-dimensional BN/SDS-PAGE analysis followed by immunoblotting with antibodies directed against NDUFS3 and ND1 (Figure 3). This figure shows a decreased amount of fully-assembled complex I and an increased amount of subcomplexes in cultured fibroblasts of the patient compared with the control, suggesting a disturbed assembly or stability of complex I. Remarkably, subcomplexes 4 and 5, as previously described,24 are accumulated, indicating that subunit NDUFA10 is incorporated in complex I after the formation of subcomplex 5. This suggests that NDUFA10 will be incorporated in a late stage of complex I assembly.
The mutations in NDUFA10 result in accumulation of subcomplexes of complex I. Two-dimensional BN/SDS-PAGE analysis followed by immunoblotting with antibodies directed against NDUFS3 to investigate the early steps of the complex I assembly, and ND1 to examine the incorporation of the mtDNA encoded subunits, shows the accumulation of mainly the subcomplexes 4 and 5 in the index patient. The subcomplexes are numbered according the assembly model described by Vogel and colleagues.24 Abbreviations: C, control; P-A10, patient with NDUFA10 mutations; C I, complex I.
Discussion
This is the first report describing compound heterozygosity for two mutations in the NDUFA10 gene in a patient with complex I deficiency, expressed in both cultured fibroblasts as well as in muscle tissue. One of the mutations is located in the sequence encoding start codon for translation and the other mutation introduced an amino acid substitution from glutamine to arginine. Several lines of evidence suggest that these compound heterozygous mutations are the underlying causes of the complex I deficiency. First, the start codon disappeared and the second mutation affected a highly conserved amino acid. Second, the father was carrier of the first mutation and the mother of the second mutation. Third, both mutations were not present in 108 controls and 21 patients with isolated complex I deficiency. Fourth, the patient fibroblasts showed decreased amount and activity of complex I, and finally, we demonstrated a disturbed assembly of complex I in the patient fibroblasts.
Besides the severely reduced complex I deficiency, the activity of complex III in muscle tissue was also somewhat reduced. This observation has been described before in several complex I deficiencies, and could be because of the fact that in the normal situation, supercomplexes can be found, in which complexes I and III occur together (CI/CIII2).25, 26, 27, 28 Therefore, screening of NDUFA10 should not be limited to only patients with isolated complex I deficiencies but also patients with a reduced complex I deficiency combined with a reduction in complex III activity should be screened for the presence of disease-causing mutations.
The seven mtDNA encoded subunits together with seven nuclear-encoded subunits form the minimal constitution of complex I, as these are involved in catalysis of electron transfer from NADH to ubiquinone and for the generation of the proton gradient. Most mutations causing complex I deficiencies have been described in these genes. However, genes encoding accessory subunits, such as NDUFS4, NDUFS6, NDUFA1, NDUFA2, and NDUFA11, may also contain pathogenic mutations.8, 9, 10, 29, 30 NDUFA10 also belongs to these accessory subunits.
Not much is known about the function of the accessory subunits, but the mutations observed in the NDUFA10 gene indicate that at least this subunit is important for proper functioning of complex I. NDUFA10 is one of the subunits that can be phosphorylated, which could be important for the function of this subunit, as phosphorylation of complex I subunits has been involved in regulation of enzymatic activity and possibly affects electron transfer.17, 18 Therefore, NDUFA10 might have a role in the control of the activity of complex I through binding of NADH.31, 32 Furthermore, it has been described that unlike other protein components of the membrane arm, NDUFA10 is a relatively hydrophilic protein and is thought to be generally weakly associated with complex I.33, 34 The phosphorylation might influence the binding affinity of NDUFA10 and regulate the amount of fully-active complex. In addition, the loose association of the NDUFA10 subunit would make it more accessible to external interactions with other proteins like kinases and phosphatases.
Complex I deficiency is one of the most frequently diagnosed defects of the OXPHOS system. At present, the diagnosis is often only based on biochemical measurements of the single enzyme activities of the OXPHOS system, as the genetic cause is still unknown in many patients. Therefore, improved methods like genomic microarray screening or exome sequencing of the nuclear genes encoding subunits and assembly factors of complex I may be useful tools in the near future. Our finding of mutations in accessory subunit NDUFA10 emphasizes that disease-causing mutations are not restricted to a certain set of subunits and genetic screening should involve all subunits. Identifying new mutations causing complex I deficiency is important to improve genetic counseling and to get more insight in the function of complex I.
Accession codes
References
Brandt U : Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 2006; 75: 69–92.
Efremov RG, Baradaran R, Sazanov LA : The architecture of respiratory complex I. Nature 2010; 465: 441–445.
Smeitink J, van den Heuvel L, DiMauro S : The genetics and pathology of oxidative phosphorylation. Nat Rev Genet 2001; 2: 342–352.
Loeffen JL, Smeitink JA, Trijbels JM et al: Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat 2000; 15: 123–134.
Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA : Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis 2006; 29: 499–515.
Fernandez-Moreira D, Ugalde C, Smeets R et al: X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol 2007; 61: 73–83.
Hoefs SJ, Dieteren CE, Distelmaier F et al: NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet 2008; 82: 1306–1315.
Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O : Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol 2008; 63: 405–408.
Ogilvie I, Kennaway NG, Shoubridge EA : A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest 2005; 115: 2784–2792.
Dunning CJ, McKenzie M, Sugiana C et al: Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 2007; 26: 3227–3237.
Saada A, Edvardson S, Rapoport M et al: C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet 2008; 82: 32–38.
Pagliarini DJ, Calvo SE, Chang B et al: A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008; 134: 112–123.
Sugiana C, Pagliarini DJ, McKenzie M et al: Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am J Hum Genet 2008; 83: 468–478.
Saada A, Vogel RO, Hoefs SJ et al: Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am J Hum Genet 2009; 84: 718–727.
Calvo SE, Tucker EJ, Compton AG et al: High-throughput, pooled sequencing identified mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 2010; 42: 851–858.
Nouws J, Nijtmans L, Houten SM et al: Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab 2010; 12: 283–294.
Schulenberg B, Aggeler R, Beechem JM, Capaldi RA, Patton WF : Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J Biol Chem 2003; 278: 27251–27255.
Schilling B, Aggeler R, Schulenberg B et al: Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett 2005; 579: 2485–2490.
Smeitink J, Sengers R, Trijbels F, van den Heuvel L : Human NADH: ubiquinone oxidoreductase. J Bioenerg Biomembr 2001; 33: 259–266.
Janssen AJ, Trijbels FJ, Sengers RC et al: Measurement of the energy-generating capacity of human muscle mitochondria: diagnostic procedure and application to human pathology. Clin Chem 2006; 52: 860–871.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Calvaruso MA, Smeitink J, Nijtmans L : Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods 2008; 46: 281–287.
Nijtmans LG, Henderson NS, Holt IJ : Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 2002; 26: 327–334.
Vogel RO, Dieteren CE, van den Heuvel LP et al: Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J Biol Chem 2007; 282: 7582–7590.
Budde SM, van den Heuvel LP, Janssen AJ et al: Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem Biophys Res Commun 2000; 275: 63–68.
Hoefs SJ, Dieteren CE, Rodenburg RJ et al: Baculovirus complementation restores a novel NDUFAF2 mutation causing complex I deficiency. Hum Mutat 2009; 30: E728–E736.
Hoefs SJ, Skjeldal OH, Rodenburg RJ et al: Novel mutations in the NDUFS1 gene cause low residual activities in human complex I deficiencies. Mol Genet Metab 2010; 100: 251–256.
Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT : Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol 2007; 27: 4228–4237.
van den Heuvel L, Ruitenbeek W, Smeets R et al: Demonstration of a new pathogenic mutation in human complex I deficiency: a 5-bp duplication in the nuclear gene encoding the 18-kD (AQDQ) subunit. Am J Hum Genet 1998; 62: 262–268.
Kirby DM, Salemi R, Sugiana C et al: NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest 2004; 114: 837–845.
Yamaguchi M, Belogrudov GI, Matsuno-Yagi A, Hatefi Y : The multiple nicotinamide nucleotide-binding subunits of bovine heart mitochondrial NADH: ubiquinone oxidoreductase (complex I). Eur J Biochem 2000; 267: 329–336.
Chen S, Guillory RJ : Identification of the NADH-NAD+ transhydrogenase peptide of the mitochondrial NADH-CoQ reductase (Complex I). A photodependent labeling study utilizing arylazido-beta-alanyl NAD+. J Biol Chem 1984; 259: 5124–5131.
Finel M, Skehel JM, Albracht SP, Fearnley IM, Walker JE : Resolution of NADH:ubiquinone oxidoreductase from bovine heart mitochondria into two subcomplexes, one of which contains the redox centers of the enzyme. Biochemistry 1992; 31: 11425–11434.
Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE : Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry 2000; 39: 7229–7235.
Acknowledgements
This work was supported by the European Community's sixth Framework Program for Research, Priority 1 ‘Life sciences, genomics and biotechnology for health’, contract number LSHM-CT-2004-005260 (MITOCIRCLE).
Web Resources
The URLs for data presented herein are as follows:
Genbank, http://www.ncbi.nlm.nih.gov/genbank
SIFT, http://sift.jcvi.org
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hoefs, S., van Spronsen, F., Lenssen, E. et al. NDUFA10 mutations cause complex I deficiency in a patient with Leigh disease. Eur J Hum Genet 19, 270–274 (2011). https://doi.org/10.1038/ejhg.2010.204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.204
Keywords
This article is cited by
-
Male guanine-rich RNA sequence binding factor 1 knockout mice (Grsf1−/−) gain less body weight during adolescence and adulthood
Cell & Bioscience (2022)
-
Most mitochondrial dGTP is tightly bound to respiratory complex I through the NDUFA10 subunit
Communications Biology (2022)
-
NDUFS6 related Leigh syndrome: a case report and review of the literature
Journal of Human Genetics (2019)
-
The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera
BMC Evolutionary Biology (2017)
-
Epigenetic profiling of human brain differential DNA methylation networks in schizophrenia
BMC Medical Genomics (2016)