Abstract
Germline mutations in CDKN2A have been reported in pancreatic cancer families, but genetic counseling for pancreatic cancer risk has been limited by lack of information on CDKN2A mutation carriers outside of selected pancreatic or melanoma kindreds. Lymphocyte DNA from consecutive, unselected white non-Hispanic patients with pancreatic adenocarcinoma was used to sequence CDKN2A. Frequencies of mutations that alter the coding of p16INK4 or p14ARF were quantified overall and in subgroups. Penetrance and likelihood of carrying mutations by family history were estimated. Among 1537 cases, 9 (0.6%) carried germline mutations in CDKN2A, including three previously unreported mutations. CDKN2A mutation carriers were more likely to have a family history of pancreatic cancer (P=0.003) or melanoma (P=0.03), and a personal history of melanoma (P=0.01). Among cases who reported having a first-degree relative with pancreatic cancer or melanoma, the carrier proportions were 3.3 and 5.3%, respectively. Penetrance for mutation carriers by age 80 was calculated to be 58% for pancreatic cancer (95% confidence interval (CI) 8–86%), and 39% for melanoma (95% CI 0–80). Among cases who ever smoked cigarettes, the risk for pancreatic cancer was higher for carriers compared with non-carriers (HR 25.8, P=2.1 × 10−13), but among nonsmokers, this comparison did not reach statistical significance. Germline mutations in CDKN2A among unselected pancreatic cancer patients are uncommon, although notably penetrant, especially among smokers. Carriers of germline mutations of CDKN2A should be counseled to avoid tobacco use to decrease risk of pancreatic cancer in addition to taking measures to decrease melanoma risk.
Similar content being viewed by others
Introduction
Pancreatic cancer is associated with very poor survival rates, with long-term survivors limited to those with resected early stage tumors. However, detection of pancreatic cancer at an early stage is challenging, and this mandates identification of high-risk groups to be targeted for prevention and screening intervention.
Since first observed by Lynch and Krush in 1968,1 the incidence of pancreatic cancer has been noted to be increased in many families affected by inherited melanoma syndromes. In genetic analyses of these families, 20–40% of inherited increased melanoma susceptibility can be linked to mutations of the CDKN2A gene (p16/Ink4) located on chromosome 9p21.2, 3, 4, 5 As a result of the increased incidence of pancreatic cancer in melanoma families with CDKN2A mutations, it was hypothesized that mutations likely also predispose to pancreatic cancer development.2 Mutations in CDKN2A have subsequently been described in familial pancreatic cancer kindreds, some without melanoma.3, 4 Somatic mutations of CDKN2A are present in up to 95% of pancreatic tumors.5 These findings provide further support for the premise that CDKN2A mutations have an important role in the development of pancreatic cancer. Clinical findings in mutation carriers clearly indicate that individuals are at higher risk for melanoma and pancreatic cancer.2 Some studies have reported associations of hypermethylation of CDKN2A6 or the A148T polymorphism in association with breast cancer risk as well.7 Brain, head/neck and non-melanoma skin cancer have also been reported to be more common among mutation carriers.8, 9, 10
The CDKN2A gene encodes two different proteins, read from alternate reading frames of a common second exon. The portion of the gene that encodes p16 is composed of three exons, and p16 is a recognized tumor suppressor because of its role in preventing progression through the G1 cell cycle checkpoint.11 It does so by preventing phosphorylation of the retinoblastoma protein, which effects a downstream inhibition of the E2F transcription factor. The other protein encoded by the CDKN2A locus, p14ARF, has negative regulatory effects on growth as it serves to stabilize p53. As p53 is activated, it interacts with various downstream targets that can arrest cyclin-dependent kinases at G1 and G2 checkpoints as well as initiate apoptosis. Both p16- and p14-mediated growth arrests appear to be involved in preventing neoplastic transformation.
There have been several difficulties inherent to studying germline CDKN2A mutations in relation to pancreatic cancer. Owing to the relatively low incidence and low survival rates, it is difficult to perform studies that have sufficient power and appropriate follow-up to determine the role of this gene in tumorigenesis. Also, the mutations have most commonly been described in families in which the predominant lesion is cutaneous malignant melanoma, with secondary attention to the pancreatic neoplasm, from which the data have been imputed. To our knowledge, there have been two studies of direct analyses for CDKN2A mutations, but in samples of <100 sporadic pancreatic cancer patients,12, 13 and none of a large, unselected sample. In addition, there has been a relative deficiency of data to estimate penetrance for known carriers of mutations predisposing to pancreatic cancer, with published data from selected melanoma kindreds.8, 14 Therefore, we performed a study in an unselected series of pancreatic adenocarcinoma patients from the Mayo Clinic Biospecimen Resource for Pancreas Research.
Patients and methods
Patient recruitment
This study was reviewed and approved by the Mayo Clinic Institutional Review Board and all patients provided written, informed consent. Pancreatic cancer patients were rapidly and systematically identified and approached, using methodology reported previously15 at Mayo Clinic Rochester, Mayo Clinic Arizona or Mayo Clinic Florida between 1 October 2000 and 1 January 2009. Of 2714 adenocarcinoma patients who were identified during this time period, 1898 consented to participate (70%), and 1605 provided blood samples for analysis, all of which were suitable for analysis. Analyses were limited to 1537 non-Hispanic whites because of the inclusion of personal and family melanoma histories, and because published mutation information is nearly exclusively generated from white subjects. Only histologically (96%) or clinically (4%) confirmed adenocarcinoma cases were included. Clinically confirmed cases required a pancreatic mass on imaging consistent with adenocarcinoma, and symptoms typical of pancreatic adenocarcinoma (weight loss, abdominal pain and painless jaundice). All cases were reviewed by subspecialist physicians with expertise in pancreatic cancer (oncologist or surgeon) for coding as adenocarcinoma. At the time of enrollment, participants were asked to complete risk factor questionnaires, including personal medical history, lifestyle behaviors, family history of cancer (including age at diagnosis) and smoking status of relatives. When risk factor questionnaire information was not completed (N=401), the medical record was abstracted to ascertain race and personal or family history of cancers. We have previously reported an intermethod reliability study in which family history of pancreatic cancer from 25 cases and 25 healthy controls was obtained from both questionnaires and abstracted from the medical record. A high degree of intermethod reliability was noted for family history (Pearson r=1.0) and race/ethnicity (1.0).15
Sequencing
Sequencing of the four exons of cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, including three exons of CDKN2A isoform 1 (NM_000077) and exon 1 of CDKN2A isoform 4 (NM_058195) was performed on subjects using dye termination chemistry (Big Dye Terminator with the model 3730 × l sequencer; Applied Biosystems, Carlsbad, CA, USA). Primer sets for polymerase chain reactions (PCRs) were designed using the web-based design tool Primer3 version 0.4.0 (http://frodo.wi.mit.edu). Intronic primers covering sequences of interest were designed at least 30 bp away from the intron/exon boundaries. PCR reactions were carried out using AmpliTaq Gold DNA Polymerase (Applied Biosystems) based on the standard protocol. After PCR reactions, the amplicons were treated with the ExoSAP-IT (USB Corp., Cleveland, OH, USA) to degrade unincorporated PCR primers and deoxynucleotide triphosphates. The cleaned products were mixed with five picomoles of the forward or reverse PCR primers for sequencing. DNA sequence variants were identified using PolyPhred.16
- CDKN2A isoform 1–exon1f:
-
5′-CAACCTGGGGCGACTTC-3′
- CDKN2A isoform 1–exon1r:
-
5′-CTGCAAACTTCGTCCTCCAG-3′
- CDKN2A isoform 1–exon2f:
-
5′-AGCTTCCTTTCCGTCATGC-3′
- CDKN2A isoform 1–exon2r:
-
5′-GCTGGAAAATGAATGCTCTG-3′
- CDKN2A isoform 1–exon3f:
-
5′-TGTGCCACACATCTTTGACC-3′
- CDKN2A isoform 1–exon3r:
-
5′-TCAGAAACGATGCTGTCTTCC-3′
- CDKN2A isoform 4–exon1f:
-
5′-CTCAGGGAAGGCGGGTG-3′
- CDKN2A isoform 4–exon1r:
-
5′-TTTTCGAGGGCCTTTCCTAC-3′
Statistical analysis
Patients were categorized by whether or not they were a carrier of a deleterious mutation in CDKN2A. These groups were then analyzed for differences in demographic characteristics using t-tests for quantitative variables and either χ2 or Fisher's exact tests for qualitative variables, depending on the sample size; relative risk estimates and 95% confidence intervals (CIs) were also calculated.17 T-tests or Wilcoxon rank-sum statistics were used to test for differences between mutation carrier status for quantitative variables. Penetrance (cumulative risk in CDKN2A mutation carriers) and 95% CIs were estimated using the kin-cohort method,18 which uses proband-reported information on first-degree relatives to estimate and compare the age-specific penetrance. First-degree relatives of mutation carriers were estimated to have a 50% mutation carrier rate, with an assumption of a 0.01% carrier rate in the general population. Ever smokers were defined as persons smoking ≥100 cigarettes in their lifetime. Cumulative risk estimates were made separately for first-degree relatives of carriers and non-carriers of CDKN2A mutations, with the baseline value for comparisons based on non-carrier relatives. Bootstrap methods were employed in the CI calculations to account for the small number of carriers. All statistical tests were two-sided and considered significant at the =0.05 level; analyses were performed in SAS version 9.1 (SAS Institute, Cary, NC, USA, http://www.sas.com) and R version 2.9.0 (R Foundation for Statistical Computing, Vienna, Austria, www.R-project.org). Only cases who completed risk factor questionnaires were included in the penetrance estimates.
Results
Mutations in p16/p14ARF
Clinical characteristics of the 1537 non-Hispanic white pancreatic cancer patients included in the analysis are shown in Table 1. The sample is 56% male with a mean age of pancreatic cancer diagnosis of 65.5±10.8 years, range 28–91. Nonsmokers comprised 41% of the sample, and first-degree family history of pancreatic cancer and melanoma were reported in 8 and 3%, respectively. Not shown in Table 1 is the residence distribution of patients: 60% of patients were residents of Minnesota, Iowa, Wisconsin, South Dakota and North Dakota, only 18 patients were non-United States (US) residents, and the remainder lived in other parts of the US.
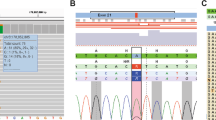
Nine (0.6%) patients were found to have a germline mutation in CDKN2A; two of the mutations alter the sequence only in the p14ARF. We identified three previously unreported mutations, including a missense mutation in p14 (A120P) and two frameshift mutations that affect both p16 and p14ARF (R80fs/P135fs and V95fs/G150fs). Demographic characteristics of mutation carriers and non-carriers are shown in Table 1, and detailed information on genetic alterations are shown in Table 2. There were no differences in sex distribution among mutation carriers vs non-carriers (P=0.74), age at diagnosis of pancreatic cancer (P=0.40), stage at presentation (P=0.41), nor location of primary tumor (P=0.30).
Of 120 cases with a first-degree relative affected with pancreatic cancer (Table 1), 4 (3.3%) carried mutations, which was a significantly higher frequency than in those without a family history (odds ratio (OR) 9.7, 95% CI 2.6–36.6, P=0.003). However, five of the nine carriers (56%) of detected mutations did not report having a family history of pancreatic cancer. Among 38 patients with melanoma in a first-degree relative, 2 (5.3%) were identified as mutation carriers, which also was higher than in those without such a family history (OR 9.1, 95% CI 1.8–45.3, P=0.03). Among 26 patients with a personal history of melanoma in addition to their pancreatic cancer, 2 (7.7%) were determined to be carriers of mutations, which is also a higher proportion than those without such a personal history (OR 17.2, 95% CI 3.4–87.0, P=0.01). Interestingly, the two cases that reported family history of melanoma were the same as the two reporting a personal history.
Estimates of CDKN2A carrier rates by personal and family history
To inform genetic counseling for pancreatic cancer risk, we examined associations of personal and family history of cancer with mutation status in the subset of cases (N=1136) who completed the risk factor questionnaire (Table 3). In this analysis, self-report of a family history of pancreatic cancer without melanoma was associated with a carrier proportion of 3.2% (3/95). No mutations were detected in cases who reported only a personal history of melanoma (N=20) or a family history of melanoma (N=26). However, in cases who reported a personal and family history of melanoma (N=2), both carried mutations in CDKN2A.
Penetrance
Cumulative risk (ie, penetrance) estimates were performed using data on first-degree relatives (N=6671) of pancreatic cancer patients who completed risk factor questionnaires that asked for detailed family history of cancer and ever/never smoking information on relatives. All nine carriers of CDKN2A mutations completed a risk factor questionnaire, providing information on their 59 first-degree relatives. Penetrance of CDKN2A mutations as manifested by incident melanoma and/or pancreatic cancer in first-degree relatives of carriers was estimated (Table 4). In mutation carriers, pancreatic cancer risk was higher among carriers than non-carriers with cumulative risk of pancreatic cancer estimated to be 57.6% by age 80 (95% CI 7.8–85.7%), in contrast to 3.2% (95% CI 2.6–4.2%) in non-carriers (log rank P=2.8 × 10−10). Similarly, melanoma was diagnosed in 39% (95% CI 0–80.2%) of carriers by age 80, in contrast to 1.1% (95% CI 0.5–1.3) of non-carriers (P=4.8 × 10−13). Interestingly, when cumulative risk estimates were limited to ever smokers, there was a significant difference in pancreatic cancer risk for carriers vs non-carriers (hazard ratio (HR) 25.8, log rank P=2.1 × 10−13). However, there was no significant difference detected among nonsmokers (HR 4.9, P=0.27). When pancreatic cancer risk was compared between ever and never smoking relatives of mutation carriers, ever-smokers appeared to have higher penetrance—92.7% (95% CI 32.6–97.9%) vs 2.6% (2.0–52.3%) by age 80, although this was not statistically significant (P=0.17) possibly because of low total numbers of cases in the comparison groups. There were 4 cases among 30 smoking relatives, and 1 among 27 nonsmoking relatives (and 1 additional case among the 2 relatives on whom no smoking information was reported).
Variants of unclear significance
Table 5 lists variants that we determined as not deleterious. Sequence variants that were detected but were of unclear significance and not included in the analysis were exon 1B (K26K, A32A, F64F), exon 1A –191A>G, –33G>C, –30G>A, –14C>T19, 20, A148T), and exon 3 (29C>G, 69C>T, 112A>G, 204T>A, 277–278insGA, 363A>G, 373C>G, 436A>G). None of these are predicted to affect initiation or splicing, nor to change the amino acid sequence except A148T (n=70), a well-documented polymorphism in 2–6% of control subjects that has not been consistently associated with melanoma risk.21, 22, 23, 24 However, we recognize the possibility that these variants could disrupt p16 or p14 expression by influencing such events as RNA splicing, mRNA stability or the translational efficacy of the mRNA.
Discussion
Our study found that the pattern and location of mutations in CDKN2A does not appear to differ significantly from that seen in melanoma kindreds,25, 26 although the frequency of mutations detected in pancreatic cancer probands (0.6%) is less than that seen in a population-based series of melanoma patients (1.8%). This is also substantially lower than the 4% frequency reported in 120 unselected Italian pancreatic cancer patients, although a founder mutation at G101W may have elevated this frequency.27 In addition to previously described mutations that affect p16 protein coding, we identified two variants that change an amino acid base in p14 but not p16 (L64L/A120P and V106V/A162T). The former has not been previously reported, while the latter has been reported at least once in a melanoma patient.28
It is difficult to know the significance of the variants apparently affecting p14ARF alone except that both patients had young-onset disease (ages 58 and 45), while neither carried additional family histories of pancreatic cancer or melanoma. However, their presence lends further evidence to the question of the significance of p14ARF in inherited risk for pancreatic cancer. Previous reports have suggested p14ARF mutations are important in pancreatic cancer risk, as in one report, two deletions in exon 1B were identified among 66 sporadic pancreatic cancer patients, but none were found among 49 familial melanoma patients.13 The GenoMEL report of CDKN2A screening in 466 melanoma families reported a higher frequency of mutations affecting both p16 and p14ARF (49%), rather than p16 alone (26%) in families with members affected with pancreatic cancer.25 However, one smaller study of 23 families with familial pancreatic cancer revealed two truncating mutations (both in families also affected by melanoma) affecting p16 but none in p14ARF.3 With regard to the type of mutation in CDKN2A, one previous report suggests that splicing mutations were more common in melanoma families affected with pancreatic cancer than in those without (17 vs 5% of mutations),29 however, the underlying reason for an individual developing pancreatic cancer rather than melanoma is not yet fully understood.
The detected exon 1A mutations L16R and R24P are known to occur in melanoma prone families.25 The D153spl mutation in the last nucleotide of exon 2 has also been previously reported,25 and affects splicing in both p16 and p14.30 Interestingly, one of the two identified carriers had a personal and family history of melanoma, while the other had only pancreatic cancer. The two frameshift mutations (R80fs and V95fs) likely substantially impact protein function because of the effect on all coding downstream of their location. They both are apparently novel, with the former coding for a hybrid p16/p14 protein after the frameshift. One of these patients reported a family history of pancreatic cancer, but no melanoma was reported in either family. Finally, a mutation detected in the 5′UTR, –34G>T, has been previously reported in melanoma kindreds,25 and creates a premature start site for translation, thereby decreasing translation of the native protein.31 The patient carrying this mutation reported a family history of pancreatic cancer.
The polymorphism A148T has been inconsistently associated with melanoma risk, with increased risk for melanoma reported in Poland (OR 2.53),32 but not France, Germany or Iceland.20, 21, 24 It has not been well studied in pancreatic cancer. We detected an allele frequency of 3.1%, similar to that of other null studies,20 although we did not test a control group. It is impossible, therefore, to draw conclusions, although the low allele frequency is suggestive that there is at most a modest association.
CDKN2A is established as a causative gene in familial pancreatic cancer families.3, 33, 34 This study represents the largest study of this gene in an unselected series of pancreatic cancer patients. As such, there are multiple observations in this descriptive study. First, there is <1% prevalence of germline CDKN2A mutations among pancreatic cancer patients. Clearly, all pancreatic cancer probands should not be tested for CDKN2A mutations. Having a family history of melanoma or pancreatic cancer, or a personal history of melanoma, however, increases the likelihood of carrying a mutation. These findings are not surprising given the existing knowledge of CDKN2A affected families, although it is also notable that mutations are only prevalent in a minority of these selected patients in our study.
Limitations of this study include the overall low number of mutations detected, which limits the power of penetrance estimates. We also were not able to directly confirm many cases of pancreatic cancer or melanoma in families—only 33% of the pancreas cancer and 23% of melanoma have been confirmed by self-report, medical records, death certificate or pathology. Smoking status for relatives was also only able to be determined by report of the proband in the majority of cases, as well. Strengths include the size of the study, the unselected nature of case ascertainment among pancreatic cancer probands, and the availability of electronic records on all patients.
Our findings may be useful for informing genetic counseling in addition to other available sources in the literature, with risk estimates of mutation detection for given scenarios in various combinations of personal and family history of melanoma and pancreatic cancer (Table 3). Our numbers for some groups are very small, and therefore the estimates are likely imprecise, although we believe they provide previously lacking information for families outside of the setting of families ascertained primarily for familial melanoma. For instance, in this study, patients who report a personal history of melanoma and pancreatic cancer had a 9.1% (2/22) (95% CI 0–22.1%) probability of harboring a mutation in CDKN2A. However, the two mutation carriers also had family histories of melanoma, while the 20 patients with melanoma and pancreatic cancer without family histories of either cancer did not carry mutations in CDKN2A. Among patients reporting a personal and family history of pancreatic cancer, 4.2% carried mutations in CDKN2A, suggesting that families carrying mutations in this gene constitute only a small minority of familial pancreatic cancer.
We have not definitively answered the question as to whether CDKN2A mutation carriers have a younger age of onset of pancreatic cancer, as our mutation carriers were slightly younger than non-carriers (mean 61.3 vs 65.6 years), and the difference did not reach statistical significance (P=0.40). At least one other study also found no difference in age at diagnosis.35 If age is at all younger among carriers, the effect is modest. We also limited our report to non-Hispanic white patients. It will be helpful in future studies to extend these analyses to other populations to determine whether the estimates we have derived can be applied to patients of all races.
The penetrance of pancreatic cancer in CDKN2A carriers—nearly 60% by age 80 (95% CI 8–86%)—is higher than previous estimates of 15–25% in melanoma family studies.8, 14 There are several potential explanations for this difference. A reporting bias could exist (over reporting in pancreatic kindreds or underreporting in melanoma kindreds), and our study is limited by relying on proband report of cancer among relatives. There also may simply be variation in our study sample compared with others, especially with regard to environmental risks (eg, high frequency of smoking). Indeed the difference in risk for carriers was only detected among ever-smokers. However, it is also possible that specific genetic differences in either the CDKN2A locus or a modifier gene could be associated with differences in penetrance as manifested by pancreatic cancer. To our knowledge, this is the first study to report penetrance that incorporates smoking status; this information is relevant from a genetic counseling standpoint, because it is a potentially modifiable risk factor. As there are a small number of cases in families of carriers, more in-depth studies could not be performed, such as whether smoking cessation altered pancreatic cancer risk. However, the difference among ever-smokers coupled with the high lifetime risk is sufficient in our judgment to promote smoking cessation/avoidance recommendations among CDKN2A carriers.
Published recommendations for CDKN2A screening include patients with multiple (≥3) primary melanomas, or families with at least one melanoma and two other instances of melanoma or pancreatic cancer in the family, with mutation detection rates of 20–40% in this setting.36, 37, 38 Under these recommendations, two of the seven (29%) families meeting criteria were determined to carry a mutation in this study, and we believe this is a reasonable proportion for discriminating candidates for genetic testing. However, it should also be noted that the majority of mutations would not have been identified with this approach. The role of clinical genetic testing for pancreatic cancer risk is still controversial in the absence of proven screening or prevention methods. However, tobacco avoidance/cessation is likely justified based on its well-established impact on pancreatic cancer risk,39, 40 in addition to the commonly recommended regular skin examinations and sun avoidance to lower melanoma risk.37
Conclusions
Germline mutations of CDKN2A among patients with pancreatic cancer are rare. Those carrying such mutations are more likely to report a personal or family history of melanoma, and a family history of pancreatic cancer. Likelihood of mutation detection by personal and family history of melanoma and pancreatic cancer are reported for genetic counseling purposes. Age at diagnosis of pancreatic cancer may be slightly younger for mutation carriers, although it did not reach statistical significance. Penetrance of pancreatic cancer and melanoma was increased among mutation carriers, with pancreatic cancer risk estimates of 58% (95% CI 8–86%) by age 80 and melanoma risk of 39% (95% CI 0–80) by age 80 in mutation carriers. Penetrance for carriers was higher among ever-smokers. Carriers of germline mutations in CDKN2A should therefore avoid tobacco use, and should be targeted for prevention and screening studies.
References
Lynch HT, Krush AJ : Heredity and malignant melanoma: implications for early cancer detection. Can Med Assoc J 1968; 99: 17–21.
Goldstein AM, Fraser MC, Struewing JP et al: Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. New Engl J Med 1995; 333: 970–974.
Bartsch DK, Sina-Frey M, Lang S et al: CDKN2A germline mutations in familial pancreatic cancer. Ann Surg 2002; 236: 730–737.
Lynch HT, Brand RE, Hogg D et al: Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: the familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer 2002; 94: 84–96.
Schutte M, Hruban RH, Geradts J et al: Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res 1997; 57: 3126–3130.
Jing F, Zhang J, Tao J et al: Hypermethylation of tumor suppressor genes BRCA1, p16 and 14–3–3sigma in serum of sporadic breast cancer patients. Onkologie 2007; 30: 14–19.
Debniak T, Gorski B, Huzarski T et al: A common variant of CDKN2A (p16) predisposes to breast cancer. J Med Genet 2005; 42: 763–765.
de Snoo FA, Bishop DT, Bergman W et al: Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res 2008; 14: 7151–7157.
Bahuau M, Vidaud D, Jenkins RB et al: Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res 1998; 58: 2298–2303.
Yarbrough WG, Aprelikova O, Pei H, Olshan AF, Liu ET : Familial tumor syndrome associated with a germline nonfunctional p16INK4a allele. J Natl Cancer Inst 1996; 88: 1489–1491.
Lukas J, Parry D, Aagaard L et al: Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 1995; 375: 503–506.
Debniak Ta, van de Wetering Ta, Scott Re et al: Low prevalence of CDKN2A/ARF mutations among early-onset cancers of breast, pancreas and malignant melanoma in Poland. Eur J Cancer Prev 2008; 17: 389–391.
Ghiorzo P, Pastorino L, Bonelli L et al: INK4/ARF germline alterations in pancreatic cancer patients. Ann Oncol 2004; 15: 70–78.
Vasen HFA, Gruis NA, Frants RR, van der Velden PA, Hille ETM, Bergman W : Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int J Cancer 2000; 87: 809–811.
McWilliams RR, Bamlet WR, de Andrade M, Rider DN, Cunningham JM, Petersen GM : Nucleotide excision repair pathway polymorphisms and pancreatic cancer risk: evidence for role of MMS19L. Cancer Epidemiol Biomarkers Prev 2009; 18: 1295–1302.
Nickerson DA, Tobe VO, Taylor SL : PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 1997; 25: 2745–2751.
Agresti A : Categorical Data Analysis, 2nd edn, John Wiley & Sons: Hoboken, NJ, 2002.
Chatterjee N, Wacholder S : A marginal likelihood approach for estimating penetrance from kin-cohort designs. Biometrics 2001; 57: 245–252.
Hashemi J, Platz A, Ueno T, Stierner U, Ringborg U, Hansson J : CDKN2A germ-line mutations in individuals with multiple cutaneous melanomas. Cancer Res 2000; 60: 6864–6867.
Goldstein AM, Stacey SN, Olafsson JH et al: CDKN2A mutations and melanoma risk in the Icelandic population. J Med Gen 2008; 45: 284–289.
Spica T, Portela M, Gerard B et al: The A148T variant of the CDKN2A gene is not associated with melanoma risk in the French and Italian populations. J Invest Dermatol 2006; 126: 1657–1660.
Hussussian CJ, Struewing JP, Goldstein AM et al: Germline p16 mutations in familial melanoma. Nat Genet 1994; 8: 15–21.
Kamb A, Shattuck-Eidens D, Eeles R et al: Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet 1994; 8: 23–26.
Pjanova D, Engele L, Randerson-Moor JA et al: CDKN2A and CDK4 variants in Latvian melanoma patients: analysis of a clinic-based population. Melanoma Res 2007; 17: 185–191.
Goldstein AM, Chan M, Harland M et al: High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006; 66: 9818–9828.
Begg CB, Orlow I, Hummer AJ et al: Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 2005; 97: 1507–1515.
Ghiorzo P, Gargiulo S, Nasti S et al: Predicting the risk of pancreatic cancer: on CDKN2A mutations in the melanoma-pancreatic cancer syndrome in Italy. J Clin Oncol 2007; 25: 5336–5337.
Orlow I, Roy P, Barz A, Canchola R, Song Y, Berwick M : Validation of denaturing high performance liquid chromatography as a rapid detection method for the identification of human INK4A gene mutations. J Mol Diagn 2001; 3: 158–163.
Goldstein AM : Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat 2004; 23: 630.
Rutter JL, Goldstein AM, Davila MR, Tucker MA, Struewing JP : CDKN2A point mutations D153spl(c.457G>T) and IVS2+1G>T result in aberrant splice products affecting both p16INK4a and p14ARF. Oncogene 2003; 22: 4444–4448.
Liu L, Dilworth D, Gao L et al: Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet 1999; 21: 128–132.
Debniak T, Scott RJ, Huzarski T et al: CDKN2A common variants and their association with melanoma risk: a population-based study. Cancer Res 2005; 65: 835–839.
Lynch HT, Brand RE, Hogg D et al: Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: the familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer 2002; 94: 84–96.
Borg A, Sandberg T, Nilsson K et al: High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst 2000; 92: 1260–1266.
Goldstein AM, Struewing JP, Fraser MC, Smith MW, Tucker MA : Prospective risk of cancer in CDKN2A germline mutation carriers. J Med Genet 2004; 41: 421–424.
Leachman SA, Carucci J, Kohlmann W et al: Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad of Dermatol 2009; 61: 677.e1–677.e14.
Hansen CB, Wadge LM, Lowstuter K, Boucher K, Leachman SA : Clinical germline genetic testing for melanoma. Lancet Oncol 2004; 5: 314–319.
Mantelli M, Barile M, Ciotti P et al: High prevalence of the G101W germline mutation in the CDKN2A (P16/ink4a) gene in 62 Italian malignant melanoma families. Am J Med Genet 2002; 107: 214–221.
Silverman DT, Dunn JA, Hoover RN et al: Cigarette smoking and pancreas cancer: a case-control study based on direct interviews. J Natl Cancer Inst 1994; 86: 1510–1516.
Fuchs CS, Colditz GA, Stampfer MJ et al: A prospective study of cigarette smoking and the risk of pancreatic cancer. Arch Intern Med 1996; 156: 2255–2260.
Acknowledgements
We thank the patients and families who participated in the Mayo Clinic Biospecimen Resource for Pancreas Research, and Traci Hammer, Jodie Cogswell, Cindy Wong, Patrick Burch, MD, Bridget Eversman, William Bamlet, Que Luu, Dennis Robinson, Martha Matsumoto, and KT Vu for their contributions to this study. This work was funded by NCI grants SPORE P50CA102701, K07116303 and R01CA97075.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
McWilliams, R., Wieben, E., Rabe, K. et al. Prevalence of CDKN2A mutations in pancreatic cancer patients: implications for genetic counseling. Eur J Hum Genet 19, 472–478 (2011). https://doi.org/10.1038/ejhg.2010.198
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.198
Keywords
This article is cited by
-
Genetic and other risk factors for pancreatic ductal adenocarcinoma (PDAC)
Familial Cancer (2024)
-
Significant detection of new germline pathogenic variants in Australian Pancreatic Cancer Screening Program participants
Hereditary Cancer in Clinical Practice (2021)
-
CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition
Hereditary Cancer in Clinical Practice (2021)
-
PARP inhibitors: shifting the paradigm in the treatment of pancreatic cancer
Medical Oncology (2021)
-
Models of pancreatic ductal adenocarcinoma
Cancer and Metastasis Reviews (2021)