Abstract
Classical lissencephaly, or isolated lissencephaly sequence (ILS), and subcortical band heterotopia (SBH) are neuronal migration disorders associated with severe mental retardation and epilepsy. Abnormalities of the LIS1 and DCX genes are implicated in the majority of patients with these disorders and account for approximately 75% of patients with ILS, whereas mutations of DCX account for 85% of patients with SBH. The molecular basis of disease in patients with ILS and SBH, in whom no abnormalities have been identified, has been questioned. We studied a series of 83 patients with ILS, SBH or pachygyria, in whom no abnormalities of the LIS1 or DCX genes had been identified, for intragenic deletions and duplications by multiplex ligation-dependent probe amplification (MLPA). In 52 patients with ILS, we identified 12 deletions and 6 duplications involving the LIS1 gene (35%), with the majority resulting in grade 3 lissencephaly. Three deletions of the DCX gene were identified in the group of nine female patients with SBH (out of 31 patients with DCX-suggestive brain anomalies), ie 33%. We estimate an overall mutation detection rate of approximately 85% by LIS1 and DCX sequencing and MLPA in ILS, and 90% by DCX sequencing and MLPA in SBH. Our results show that intragenic deletions and duplications of the LIS1 and DCX genes account for a significant number of patients with ILS and SBH, where no molecular defect had previously been identified. Incorporation of deletion/duplication analysis of the LIS1 and DCX genes will be important for the molecular diagnosis of patients with ILS and SBH.
Similar content being viewed by others
Introduction
Classic lissencephaly (LIS) and subcortical band heterotopia (SBH) comprise a spectrum of severe cortical malformations resulting from abnormal neuronal migration during brain development. The most severe form of LIS consists of absent gyri (or agyria), while a less severe or intermediate form consists of abnormally wide gyri (or pachygyria).1, 2, 3 The less severe forms of LIS merge with the phenotype of SBH, a less severe cortical malformation characterized by the presence of bilateral smooth bands of gray matter in the central or superficial white matter of the brain.4 Both LIS and SBH may occur in isolated form as isolated lissencephaly sequence (ILS) or isolated SBH, or as part of malformation syndromes such as Miller–Dieker or Baraitser–Winter syndrome, with the former comprising a contiguous gene deletion disorder resulting from large microdeletions of the 17p13.3 region.5, 6, 7, 8
The genetic basis of ILS and SBH are reasonably well understood, with specific patterns of brain malformation emerging that can assist in distinguishing the gene in which a causative mutation is most likely to be found, most often either the LIS1 (PAFAH1B1) or DCX gene. Abnormalities in the LIS1 gene, located on chromosome 17p13.3, cause a malformation pattern or gradient that is more severe in posterior as compared to anterior brain regions (posterior-to-anterior), while mutations in the DCX gene result in brain malformation patterns that are more severe in the frontal regions of the brain becoming less severe posteriorly (anterior-to-posterior).1, 3, 9
Abnormalities of the LIS1 and DCX genes account for approximately 75% of ILS.2, 3, 10 LIS1 abnormalities comprise whole gene deletions that are detectable by fluorescence in situ hybridization (FISH) analysis and mutations within the gene that are detectable by DNA sequencing. DCX abnormalities that result in ILS consist primarily of mutations within the gene. Mutations in DCX have been described in approximately 38% of males with ILS but only very rarely in females with ILS, and in approximately 85% of females and approximately 29% of males with SBH.2, 3, 11, 12, 13 For both the LIS1 and DCX genes, mutations that abolish function such as entire gene deletions, nonsense, frameshift and splicing mutations generally result in a more severe phenotype, while missense mutations tend to result in the milder phenotypes.9, 10, 13, 14
Intragenic deletions of the LIS1 and DCX genes also result in lissencephaly, but the frequency of these types of mutation as well as the associated disease severity has not been well characterized. Intragenic deletion mutations of the LIS1 gene, detected by Southern blot, have previously been found to account for approximately 8% of disease-causing mutations.2, 10 We set out to determine the incidence of intragenic deletions and duplications in the LIS1 and DCX genes in patients with varying degrees of lissencephaly and SBH.
In this study, we systematically screened a cohort of 83 patients for LIS1 and DCX intragenic deletions and duplications who were previously found to be negative for microdeletions in the 17p13.3 region by FISH and also negative for mutations detected by sequencing of the DCX and LIS1 genes. The cohort of patients studied has been well characterized in terms of brain abnormality patterns and disease severity. This study provides a comprehensive analysis of the DCX and LIS1 genes and allows us to correlate deletions and duplications of these two genes with disease severity. Our results indicate that deletions and duplications of the LIS1 and DCX genes account for up to 35% of patients with lissencephaly and SBH in whom the molecular basis of disease had previously not been identified. With the inclusion of deletion/duplication analysis of the LIS1 and DCX genes, the molecular basis of 85–90% of patients with lissencephaly and SBH can now be determined.
Materials and methods
Patient selection
Blood samples were obtained from patients after informed consent was given. Protocols were approved by the appropriate IRB committees before the initiation of the research.
Patients
For this study, we classified patients as having lissencephaly based on the presence of agyria, pachygyria, SBH or combinations of these malformations. We defined agyria as areas of cortex lacking sulci (convolutions or gyri) larger than 3 cm and pachygyria as areas of cortex lacking sulci between ∼1.5 and 3 cm, both with the cortical ribbon typically thicker than 1 cm. We used the same grading system as in our earlier studies, which consists of completely absent gyri or agyria (grade 1), diffuse agyria with a few shallow sulci usually frontally (grade 2), mixed agyria and abnormally broad gyri or pachygyria (grade 3), diffuse pachygyria (grade 4), mixed pachygyria and SBH (grade 5) and SBH only (grade 6).1, 3, 9, 10
We searched our large brain malformation database for all patients with ILS or SBH for whom original brain imaging studies were available and reviewed by one of the authors (WBD). All patients were classified with attention to the pattern (severity and gradient), and the presence or absence of associated features. We excluded all patients with syndromic forms of lissencephaly, including lissencephaly with moderate or severe cerebellar hypoplasia (many patients with DCX or LIS1 mutations have mild cerebellar vermis hypoplasia), Baraitser–Winter syndrome, X-linked lissencephaly with abnormal genitalia, and several other novel syndromes observed in single patients. We separated the remaining patients into three groups based on the pattern of the cortical malformation. From this group, we then selected 83 patients who met the following criteria: negative findings by FISH for a microdeletion of the 17p13.3 region and in whom no mutations were identified in either the LIS1 and/or DCX genes by sequence analysis (Supplementary Table 1).
In total, 52 patients exhibiting severe lissencephaly in the parietal and occipital regions of the brain (severity following an posterior-to-anterior gradient – ie, an LIS1 pattern) were investigated for intragenic deletions and duplications of the LIS1 gene, while 31 patients (22 females and 9 males) with SBH and lissencephaly encompassing the frontal regions of the brain (severity following an anterior-to-posterior gradient – ie, a DCX pattern) were analyzed for intragenic deletions and duplications in the DCX gene. The patient cohort consisted of 83 subjects in total.
DNA isolation
DNA was extracted from leukocytes from EDTA-treated blood using either the Puregene kit (Gentra Systems Inc., Minneapolis, MN, USA) or the MagNAPure Total Nucleic Acid Extraction system (Roche Diagnostics, Indianapolis, IN, USA) following the manufacturer's protocols.
Detection of intragenic deletions and duplications
We used multiplex ligation-dependent probe amplification (MLPA) to screen our series of 83 patients. All 83 patients had been analyzed for mutations in the LIS1 and/or DCX genes and the 17p13.3 microdeletion and found to be negative. Analysis of the LIS1 and DCX genes was performed using the Lissencephaly P061 MLPA kit (MRC-Holland, Amsterdam, the Netherlands) following the manufacturer's protocol. The MLPA kit contains probes for all the exons (coding and upstream non-coding) of the DCX gene and all exons (coding and upstream non-coding) of the LIS1 gene plus the LIS1-flanking genes, YWHAE, HIC1, MGC3329, KIAA0664 and KIAA1039. MLPA fluorescently labeled PCR products were size fractionated on the automated fragment analyzer ABI3130 (Applied Biosystems, Foster City, CA, USA) and analyzed using the fragment sizing program GeneMarker (Soft Genetics Inc.). Positive controls (samples with microdeletions of 17p13.3) and normal controls were included with each run. Ratios of 0.4–0.6 or x>1.4 were considered to denote the presence of a deletion or duplication, respectively, while a ratio of 0.8–1.2 was considered to be normal by MLPA. Any MLPA results that indicated the presence of a single or multiple exon deletion or duplication in a particular patient sample were confirmed by real time quantitative-PCR (RT-qPCR). RT-qPCR was performed using an internal control gene along with the regions of the LIS1 and DCX genes being tested. Short amplicons between 60 and 100 bases were amplified utilizing PowerSYBR Green (Applied Biosystems) following the manufacturer's protocol, and the ratio of the LIS1/DCX region of interest to internal control gene amplification was compared to normal controls. Two independent amplicons within the region containing a potential deletion or duplication were analyzed by RT-qPCR and were run concurrently in duplicate with positive and normal controls. A ratio of 0.4–0.6 represented the presence of a deletion, while a value over 1.4 was interpreted as a duplication. A ratio of 0.8–1.2 was indicative of no deletion or duplication.
Results
Intragenic deletion/duplication analysis of the LIS1 gene
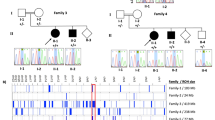
Of the 52 patients studied for LIS1 deletions/duplications, 18 patients were found to have either a deletion or duplication affecting the LIS1 gene (35%) (Table 1). Twelve of the 18 patients harbored a deletion, whereas 6 were found to have a duplication (Figure 1). One patient with a LIS1 deletion also had a duplication involving the upstream HIC1 gene. The deletions and duplications identified were of varying sizes ranging from the deletion or duplication of a single exon to deletions involving the entire coding region of the LIS1 gene. Although the deletions and duplications were found scattered within the gene, exons 3–6 appeared to be involved in the majority of cases (11 of 18). Several of the deletions included genes that flank the LIS1 gene, such as the MGC3329 and HIC1 genes upstream and the KIAA0664 and KIAA1039 genes downstream (6 of 18). Two of the deletions involved the upstream non-coding region of LIS1 and did not appear to include any of the coding region. One duplication was found to involve only the downstream KIAA0664 gene and did not appear to involve any of the coding and non-coding regions of the LIS1 gene. These results suggest the presence of regulatory elements important for LIS1 expression that are located both upstream and downstream of the gene.
Schematic of intragenic deletions and duplications detected in the LIS1 and surrounding genes. Each bar denotes a deletion/duplication identified in a patient with the deletions depicted on top and the duplications depicted below the diagram of the gene. The bars indicate the extent of the deletions/duplications and show the regions of the gene/s involved. The associated lissencephaly grade for each patient in whom a deletion/duplication was identified is also indicated. The 11 exons of the LIS1 gene are shown to be located between the genes MGC3329 and KIAA0064, with non-coding exons and untranslated portions of exons indicated in white and the coding exons indicated in gray. The circled region in the top left corner represents an individual that was found to have an intragenic deletion encompassing exons 1–6 of the LIS1 gene as well as a duplication of the HIC1 gene. This figure is not drawn to scale.
Of the 18 deletions and duplications identified, 16 were found in patients with grade 3 LIS (Table 3). Twenty of the 52 patients studied had grade 3 LIS, resulting in 80% of this group (16 of 20) harboring either a deletion or duplication. All six duplications identified were in patients with grade 3 lissencephaly, while 10 of the 12 deletions were present in this group. Of the six duplications, four involved multiple exons, one was a duplication encompassing a single exon (exon 5) and one duplication was of a flanking gene 3′ of the LIS1 gene (KIAA0664). All five duplications of the LIS1 exonic region involved the coiled-coil domain as well as one or more of the WD40 repeats, with three including the critical second WD40 repeat. The four multiple exonic duplications are predicted to be out-of-frame, while the exon 5 duplication is predicted to result in a duplication of a portion of the coiled-coil and the first WD40 domains. The sixth duplication does not appear to affect the LIS1 gene but rather involves a downstream gene, KIAA0664. We have not characterized the size of this duplication further, so it remains possible (and most likely) that 3′UTR and regulatory elements of the LIS1 gene are affected.
All 10 of the deletions seen in the grade 3 lissencephaly patients are predicted to be deleterious. Two of the deletions appeared to involve the entire coding region of the LIS1 gene as well as additional downstream genes. These deletions were not detectable by FISH analysis indicating that the size is smaller than that traditionally detected by this technology. Of the smaller deletions that involved a part of the LIS1 gene, three included exon 2, which harbors the ATG translational start site. Three of the intragenic deletions involved multiple or single exons that were all predicted to alter the reading frame and truncate the protein. The remaining two deletions included the non-coding exon 1 of the LIS1 gene and additional upstream genes. These deletions are predicted to affect promoter and regulatory elements of the LIS1 gene. A significant number of the intragenic deletion and duplication break points were located within introns 1, 2 and 5 (44.8%).
Of 22 patients with grade 4 lissencephaly, 2 were identified as having an intragenic deletion of the LIS1 gene, ie 9%. Both were single exon deletions (exons 5 and 9) and resulted in in-frame deletions within the LIS1 gene. The complete deletion of exon 5 or 9 results in disruption of the first WD40 repeat and the fourth and fifth WD40 repeats, respectively.3 Exon 5 also contains the coiled-coil domain (amino acids 44–78), which may play a role in PAFAH1B1 protein–protein interaction.10 The disruption of these important functional domains of the LIS1 gene most likely explains the resulting lissencephaly phenotype observed. These two deletions may represent slightly milder mutations that manifest as grade 4 lissencephaly, as they do not result in a frame shift causing a prematurely truncated PAFAH1B1 protein.
No deletions or duplications of the LIS1 gene were detected in the small group of patients with grade 1 or 2 lissencephaly (0 out of 5 patients). Our retrospective review of their phenotype showed atypical features in four of five patients, consisting of mild to borderline moderate cerebellar vermis hypoplasia in three patients, which was not quite sufficient for us to exclude them from the study, and congenital microcephaly that was more severe than usual with mutations of DCX or LIS1 (−3 standard deviations) in the other patient. In addition, no deletions or duplications of the LIS1 gene were found in the group of patients with SBH and a posterior-to-anterior gradient of severity (0 out of 5 patients). Although SBH is generally associated with defects in the DCX gene, a posterior-to-anterior grade of severity has been associated with LIS1 mutations.11, 15 This group of patients was also found not to have deletions or duplications of the DCX gene.
Intragenic deletion/duplication analysis of the DCX gene
Patients included for DCX analysis included those with either SBH, pachygyria-SBH or pachygyria manifesting with an anterior-to-posterior gradient that is suggestive of a DCX abnormality.16 This group included 22 females and 9 males (Table 2). Three deletions of the DCX gene were observed, all in female patients (Figure 2). Two of the deletions involved exons 3 and 4 and one involved exons 6 and 7 (Table 3). Exon 4 contains the translation start site and a deletion of this region is predicted to affect protein production. The deletion involving exons 6 and 7 is out-of-frame and therefore either leads to loss of the protein through the nonsense-mediate mRNA decay pathway,17 or truncates the protein leading to loss of the carboxyl end of the protein that contains a CDK5 phosphorylation site at Ser297 and a serine-proline-rich region containing at least four other potential phosphorylation sites near the end of the coding region.18, 19
Schematic of intragenic deletions detected in the DCX gene in three female patients with subcortical band heterotopia (SBH) or SBH-pachygyria. The A group indicates the DCX intragenic deletions identified in two females with SBH, while B represents the deletion identified in one female with SBH-pachygyria. The solid black bars indicate the minimum size of the intragenic deletions and show the region of the gene involved. This figure is not drawn to scale.
All three deletions occurred in the group of nine female patients with either SBH or SBH-pachygyria, ie 33%. None of the 13 female patients with the more severe pachygyria phenotype (lissencephaly grade 4) had DCX abnormalities. In addition, none of the male patients included in this group with either SBH-pachygyria or pachygyria (lissencephaly grades 4 and 5) were found to have DCX abnormalities. These results further increase the percentage of female patients with SBH and DCX abnormalities.
Discussion
To further characterize the molecular basis of the neuronal migration disorder lissencephaly and the phenotypically milder SBH, we undertook a study to examine the frequency of intragenic deletions and duplications in the LIS1 and DCX genes. Our results showed that deletions and/or duplications in both the LIS1 and DCX genes account for a significant proportion of patients with ILS and SBH.
Through systematic phenotypic classification of brain malformation gradients and severity in ILS and SBH patients negative for a 17p13.3 microdeletion detected by FISH, or a point mutation in LIS1 or DCX detected by sequencing, we were able to identify 18 intragenic deletions and duplications of the LIS1 gene in our group of 52 patients. Deletions and duplications of the LIS1 gene therefore appear to account for approximately 35% of this group. When divided according to the severity of LIS, we found that a large majority of deletions and duplications occurred in patients with the classical grade 3 form of LIS. In fact, 80% (16 of 20) of our patients in this group were found to harbor a deletion or duplication of LIS1. The milder grade 4 form of LIS (which consists of complete pachygyria) had a lower incidence of deletions and duplications with only 9% (2 of 22) of patients in this group having an intragenic deletion. No deletions or duplications were identified in patients with even milder grade 6 form of LIS (posterior SBH). These results suggest that the milder forms of LIS are genetically heterogeneous and that perhaps other genetic factors may be involved in their etiology. In addition, intragenic deletions and duplications were not found in the more severe group (grades 1 and 2 that consist of complete or nearly complete agyria). These results appear to differ somewhat from our previous findings where LIS1 deletions were identified in some patients with grade 2 LIS.10 This may be due to the small number of patients with grade 2 LIS included in this study. This group of patients often has severe DCX mutations in boys or microdeletions of the 17p13.3 region, but our experience suggests that additional causative genes are most likely to be found, such as the new TUBA1A gene.20
Intragenic deletions of the DCX gene were found in 33% of female patients with SBH in whom no mutations of DCX had previously been identified. The SBH phenotype in the patients with DCX deletions was similar to those with DCX mutations detected by sequencing. Two of the deletion patients had typical diffuse SBH, while another had a slightly more severe phenotype with a combination of pachygyria and SBH. The pachygyria-SBH phenotype showed an anterior greater than posterior severity that has been associated with DCX abnormalities.3 A series of 20 patients (13 females and 7 males) with the more severe pachygyria-only phenotype showing an anterior-to-posterior gradient were also analyzed for DCX with no deletions or duplications identified. On reviewing the phenotype of these patients, we found that most have somewhat less extensive pachygyria and less marked thickening of the cortex than typical for DCX mutations. Our entire research database shows DCX mutations in only 36 of 138 (26%) patients with frontal predominant pachygyria. We take these observations together to mean that other gene(s) are responsible for this phenotype. Our results suggest that intragenic deletions and duplications of DCX result in a more classic SBH phenotype.
The intragenic deletions and duplications identified were of varying sizes and encompassed different regions of the LIS1 and DCX genes. While there were no obvious hot spots for deletions and duplications, break points within introns 1, 2 and 5 appeared to be more common in the LIS1 gene. The intragenic deletions and duplications observed are all predicted to be loss-of-function mutations that either result in an unstable protein or the lack of protein. Interestingly, the two small in-frame deletions in the LIS1 gene were found in patients with less severe lissencephaly (grade 4) compared to the remaining out-of-frame deletions and duplications (all found in grade 3 patients). It is possible that the in-frame deletions result in protein that retain some function and manifest as a milder phenotype, as we have observed for some missense mutations of LIS1.14
The smallest deletions/duplications observed in the LIS1 gene involved only a single exon, whereas the largest appeared to involve the entire coding region plus additional flanking genes. The exact size of the deletions and duplications however was not determined. Two of the largest deletions that affected the entire LIS1 coding region have a minimum size of ∼158 kb (LIS1 exon 2 to KIAA1039 exon 1). These deletions were not detected by previous FISH analysis. Smaller deletions of the 17p13.3 region that partially affect the LIS1 gene have previously been described that were detected by LIS1-specific FISH probes but missed by the commercially available FISH probe for LIS1.21
We identified an impressive number of intragenic LIS1 duplications in our cohort of ILS patients. Of the 18 abnormalities identified, one-third were duplications that involve one or multiple exons. Three of the multiple exonic duplications included the critical second WD40 repeat, whereas the other two intragenic LIS1 gene duplications encompassed the first WD40 repeat as well as the coiled-coil domain. As the WD40 domains and protein interaction domains are considered to be extremely important for the correct folding and function of the PAFAHB1 protein, intragenic duplications could represent a severe type of mutation that causes significant disruption of the LIS1 gene product. The sixth duplication occurred downstream of the LIS1 gene and most likely affects important regulatory elements, although this has not been proven conclusively. Intragenic duplications, in general, tend to be less described as a disease-causing mechanism compared to intragenic deletions. This could be due to duplications occurring less frequently, not resulting in a recognizable phenotype, or not being detected. As the mechanisms that give rise to intragenic deletions are also predicted to result in intragenic duplications, the incidence of both should be similar and it is possible that duplications are being under-detected. Our results showed that intragenic duplications of the LIS1 gene are a significant cause of disease in ILS.
Microdeletions of 17p13.3 are found in about 40% of patients, while mutations within the LIS1 and DCX genes are seen in approximately 35% of patients.2, 3 Our results indicate that intragenic deletions and duplications of the LIS1 gene account for an additional 10% of this group (35% of patients without microdeletions or mutations). These results are similar to Mei et al (2008) who described the presence of LIS1 intragenic deletions and duplications in 42% of their ILS patients negative for a 17p13.3 microdeletion detected by FISH,22 however we identified a larger percent of intragenic duplications in our set of patients. A combination of DNA sequencing and deletion/duplication analysis of the LIS1 and/or DCX genes will therefore detect abnormalities in approximately 85% of patients with ILS, considering 17p13.3 microdeletions, LIS1 and DCX mutations detected by sequencing (which includes DCX deletions in males) and LIS1 intragenic deletions/duplications. Mutations in the TUBA1A gene have also been implicated in a small percentage of patients with LIS or SBH,20, 23 and result in a malformation pattern very similar to that caused by defects in the LIS1 gene, although additional features have been recognized that may distinguish a subset of these patients. Mutations in the other three known LIS genes – ARX, RELN and VLDLR – have been associated with additional abnormalities that exclude them from the ILS category, and none of these genes are associated with SBH.
Similarly, the molecular basis of approximately 90% of SBH is now understood. Mutations of the DCX gene are present in approximately 85% of SBH patients24 and we have found intragenic deletions of the DCX gene in an additional 4% (27% of patients without DCX mutations). Our results are similar to Mei et al (2007) who described the presence of DCX intragenic deletions in 27% of their SBH patients in whom no DCX mutations had been identified by sequencing.25 A combination of DNA sequencing and deletion/duplication analysis of the DCX gene will therefore detect abnormalities in approximately 90% of patients with SBH. The remaining 10% of SBH are most likely due to abnormalities in other genes, such as TUBA1A.
Intragenic deletions and duplications are emerging as contributing to a significant percentage of the molecular basis of disease in multiple different diseases. As these abnormalities have been well documented to contribute significantly to genetic disorders, such as Duchenne and Becker muscular dystrophies, their presence has remained under-detected. This has largely been due to the more common technologies used such as FISH and BAC comparative genomic hybridization that detect larger genomic deletions and duplications (>80 kb) and DNA sequencing that detects smaller deletions and duplications (ranging from a few base pairs to couple of hundred base pairs), all of which will miss exonic deletions and duplications. Southern blot analysis can detect this intermediate size deletion/duplication, but this technique is not routinely or widely used for mutation analysis. Technologies for intragenic deletion/duplication analysis that are more amenable to routine use such as MLPA or RT-qPCR and more recently oligo array CGH will be useful for better characterization of this group of mutations. Intragenic deletions and duplications are likely to account for a sizeable fraction of mutations in genetic disease, and comprehensive mutation analysis should therefore include DNA sequencing as well as intragenic deletion/duplication analysis.
Our findings show that the lissencephaly phenotype associated with mutations and deletions/duplications (both intragenic and large microdeletions) in the LIS1 and DCX genes are very similar. On the basis of these findings, we recommend intragenic deletion/duplication testing by MLPA, or other intragenic deletion/duplication method as the initial assessment (this will detect both intragenic deletions/duplications and large microdeletions), followed by sequencing of LIS1 and then DCX for a classic LIS phenotype. For SBH, we recommend DCX sequencing followed by MLPA. In a subset of male patients with grade 1 or frontal lissencephaly, we suggest DCX sequencing before LIS1 sequencing or deletion/duplication analysis. To perform the most appropriate range of testing, a thorough review of patient information by an experienced physician will most likely increase accuracy and mutation yield from genetic testing.
In conclusion, our study of a large group of well-characterized patients with ILS and SBH shows that intragenic deletions and duplications of the LIS1 and DCX genes play a significant role in these disorders and our findings highlight the importance of developing systematic approaches and methods for detection of such mutations. These results are important for diagnostic and genetic counseling purposes. Genetic testing for ILS and SBH should include both mutation and deletion/duplication analysis for the LIS1 and DCX genes.
References
Dobyns WB, Truwit CL, Ross ME et al: Differences in the gyral pattern distinguish chromosome 17-linked and X-linked lissencephaly. Neurology 1999; 53: 270–277.
Pilz DT, Macha ME, Precht KS, Smith AC, Dobyns WB, Ledbetter DH : Fluorescence in situ hybridization analysis with LIS1 specific probes reveals a high deletion mutation rate in isolated lissencephaly sequence. Genet Med 1998; 1: 29–33.
Pilz DT, Matsumoto N, Minnerath S et al: LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum Mol Genet 1998; 7: 2029–2037.
Barkovich AJ, Jackson Jr DE, Boyer RS : Band heterotopias: a newly recognized neuronal migration anomaly. Radiology 1989; 171: 455–458.
Cardoso C, Leventer RJ, Ward HL et al: Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet 2003; 72: 918–930.
Dobyns WB, Curry CJ, Hoyme HE, Turlington L, Ledbetter DH : Clinical and molecular diagnosis of Miller-Dieker syndrome. Am J Hum Genet 1991; 48: 584–594.
Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH : Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet 1997; 6: 157–164.
Rossi M, Guerrini R, Dobyns WB, Andria G, Winter RM : Characterization of brain malformations in the Baraitser-Winter syndrome and review of the literature. Neuropediatrics 2003; 34: 287–292.
Cardoso C, Leventer RJ, Dowling JJ et al: Clinical and molecular basis of classical lissencephaly: mutations in the LIS1 gene (PAFAH1B1). Hum Mutat 2002; 19: 4–15.
Cardoso C, Leventer RJ, Matsumoto N et al: The location and type of mutation predict malformation severity in isolated lissencephaly caused by abnormalities within the LIS1 gene. Hum Mol Genet 2000; 9: 3019–3028.
D’Agostino MD, Bernasconi A, Das S et al: Subcortical band heterotopia (SBH) in males: clinical, imaging and genetic findings in comparison with females. Brain 2002; 125: 2507–2522.
Dobyns WB, Elias ER, Newlin AC, Pagon RA, Ledbetter DH : Causal heterogeneity in isolated lissencephaly. Neurology 1992; 42: 1375–1388.
Gleeson JG, Minnerath SR, Fox JW et al: Characterization of mutations in the gene doublecortin in patients with double cortex syndrome. Ann Neurol 1999; 45: 146–153.
Leventer RJ, Cardoso C, Ledbetter DH, Dobyns WB : LIS1 missense mutations cause milder lissencephaly phenotypes including a child with normal IQ. Neurology 2001; 57: 416–422.
Sicca F, Kelemen A, Genton P et al: Mosaic mutations of the LIS1 gene cause subcortical band heterotopia. Neurology 2003; 61: 1042–1046.
Dobyns WB, Berry-Kravis E, Havernick NJ, Holden KR, Viskochil D : X-linked lissencephaly with absent corpus callosum and ambiguous genitalia. Am J Med Genet 1999; 86: 331–337.
Maquat LE, Carmichael GG : Quality control of mRNA function. Cell 2001; 104: 173–176.
Gleeson JG, Allen KM, Fox JW et al: Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell 1998; 92: 63–72.
Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG : Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron 2004; 41: 215–227.
Poirier K, Keays DA, Francis F et al: Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum Mutat 2007; 28: 1055–1064.
Izumi K, Kuratsuji G, Ikeda K, Takahashi T, Kosaki K : Partial deletion of LIS1: a pitfall in molecular diagnosis of Miller-Dieker syndrome. Pediatr Neurol 2007; 36: 258–260.
Mei D, Lewis R, Parrini E et al: High frequency of genomic deletions and duplication in the LIS1 gene in lissencephaly: implications for molecular diagnosis. J Med Genet 2008; 45: 355–361.
Keays DA, Tian G, Poirier K et al: Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 2007; 128: 45–57.
Matsumoto N, Leventer RJ, Kuc JA et al: Mutation analysis of the DCX gene and genotype/phenotype correlation in subcortical band heterotopia. Eur J Hum Genet 2001; 9: 5–12.
Mei D, Parrini E, Pasqualetti M et al: Multiplex ligation-dependent probe amplification detects DCX gene deletions in band heterotopia. Neurology 2007; 68: 446–450.
Acknowledgements
We thank the patients and their families for their participation in this study. This study was supported in part by Grants to WBD (P01-NS039404 and R01-NS058721).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Haverfield, E., Whited, A., Petras, K. et al. Intragenic deletions and duplications of the LIS1 and DCX genes: a major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur J Hum Genet 17, 911–918 (2009). https://doi.org/10.1038/ejhg.2008.213
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.213
Keywords
This article is cited by
-
Connecting DCX, COMT and FMR1 in social behavior and cognitive impairment
Behavioral and Brain Functions (2022)
-
High content screening and proteomic analysis identify a kinase inhibitor that rescues pathological phenotypes in a patient-derived model of Parkinson’s disease
npj Parkinson's Disease (2022)
-
A novel missense variant in the EML1 gene associated with bilateral ribbon-like subcortical heterotopia leads to ciliary defects
Journal of Human Genetics (2021)
-
Ultrasonic Diagnosis of Lissencephaly: Literature Review and A Case Report
Journal of Fetal Medicine (2021)
-
WD40 repeat domain proteins: a novel target class?
Nature Reviews Drug Discovery (2017)