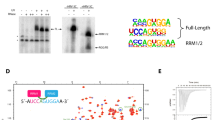
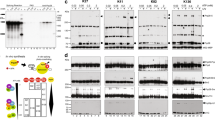
Abstract
In metazoans, spliceosome assembly is initiated through recognition of the 5′ splice site by U1 snRNP and the polypyrimidine tract by the U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor, U2AF (refs 1, 2). U2AF is a heterodimer comprising a large subunit, U2AF65, and a small subunit, U2AF35 (ref. 3). U2AF65 directly contacts the polypyrimidine tract and is required for splicing in vitro4. In comparison, the role of U2AF35 has been puzzling: U2AF35 is highly conserved5,6,7 and is required for viability6,7, but can be dispensed with for splicing in vitro4,8,9. Here we use site-specific crosslinking to show that very early during spliceosome assembly U2AF35 directly contacts the 3′ splice site. Mutational analysis and in vitro genetic selection indicate that U2AF35 has a sequence-specific RNA-binding activity that recognizes the 3′-splice-site consensus, AG/G. We show that for introns with weak polypyrimidine tracts, the U2AF35–3′-splice-site interaction is critical for U2AF binding and splicing. Our results demonstrate a new biochemical activity of U2AF35, identify the factor that initially recognizes the 3′ splice site, and explain why the AG dinucleotide is required for the first step of splicing for some but not all introns.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Reed, R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 6, 215–220 (1996).
Reed,R. & Palandjian,L. in Spliceosome Assembly In Eukaryotic mRNA Processing (ed. Krainer, A.) 103–129 (IRS Press, Oxford, 1997).
Zamore,P. & Green,M. R. Identification, purification and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA 86, 9243–9247 (1989).
Zamore,P. D., Patton,J. G. & Green,M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355, 609–614 (1992).
Wentz-Hunter,K. & Potashkin,J. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 24, 1849–1854 (1996).
Rudner,D. Z., Kanaar,R., Breger,K. S. & Rio,D. C. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl Acad. Sci. USA 93, 10333–10337 (1996).
Zorio,D. A. & Blumenthal,T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5, 487–494 (1999).
Valcarcel,J., Gaur,R. K., Singh,R. & Green,M. R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273, 1706–1709 (1996).
Kan,J. L. & Green,M. R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 15, 462–471 (1999).
Lamond,A. I., Konarska,M. M. & Sharp,P. A. A mutational analysis of spliceosome assembly: evidence for the splice site collaboration during spliceosome formation. Genes Dev. 1, 532–543 (1987).
Reed,R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3, 2113–2123 (1989).
Singh,R., Valcarcel,J. & Green,M. R. District binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 (1995).
Senapathy,P., Shapiro,M. B. & Harris,N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183, 252–278 (1990).
Mount,S. M. et al. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 20, 4255–4262 (1992).
Nilsen,T. W. in Trans-Splicing In Eukaryotic mRNA Processing (ed. Krainer, A.) 310–334 (IRS Press Oxford, 1997).
Abovich,N. & Rosbash,M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 2, 403–412 (1997).
Romfo,C. M. & Wise,J. A. Both the polypyrimidine tract and the 3′ splice site function prior to the first step of splicing in fission yeast. Nucleic Acids Res. 15, 4658–4665 (1997).
Fleckner,J., Zhang,M., Valcarcel,J. & Green,M. R. U2AF65 recruits a novel human dead box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11, 1864–1872 (1997).
Zuo,P. & Maniatis,T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10, 1356–1368 (1996).
Wu,S. & Green,M. R. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 16, 4421–4432 (1997).
Gama-Carvalho,M. et al. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137, 975–987 (1997).
Acknowledgements
We thank S. DuPont, J. Varcarcel, R. Singh, J. Kan and P. Maroney for suggestions and reagents; R. Reed for β-globin derivatives; P. Zuo and T. Maniatis for U2AF35 antibody and baculovirus U2AF35 expression construct; M. Gama-Carvalho and M. Carmo-Fonseca for U2AF65 monoclonal antibody (MC3); L. Chiang and J. Wang for nuclear extracts. We thank T. Blumenthal for communicating results before publication. This work was supported in part by grants from the NIH to M.R.G. and T.W.N. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, S., Romfo, C., Nilsen, T. et al. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835 (1999). https://doi.org/10.1038/45590
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/45590
This article is cited by
-
Impact of U2AF1 mutations on circular RNA expression in myelodysplastic neoplasms
Leukemia (2023)
-
Recruitment of a splicing factor to the nuclear lamina for its inactivation
Communications Biology (2022)
-
Emerging roles of spliceosome in cancer and immunity
Protein & Cell (2022)
-
U2af1 is required for survival and function of hematopoietic stem/progenitor cells
Leukemia (2021)
-
Complex landscape of alternative splicing in myeloid neoplasms
Leukemia (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.