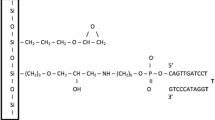
Abstract
The structural features of nucleic acid probes tethered to a solid support and the molecular basis of their interaction with targets in solution have direct implications for the hybridization process. We discuss how arrays of oligonucleotides provide powerful tools to study the molecular basis of these interactions on a scale which is impossible using conventional analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gillespie, D. & Spiegelman, S. A quantitative assay for DNA–RNA hybrids with DNA immobilised on a membrane. J. Mol. Biol. 12, 829–842 (1965).
Ritossa, F., Malva, C., Boncinelli, E., Graziani, F. & Polito, L. The first steps of magnification of DNA complementary to ribosomal RNA in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 68, 1580– 1584 (1971).
Birnstiel, M., Speirs, J., Purdom, I., Jones, K. & Loening, U.E. Properties and composition of the isolated ribosomal DNA satellite of Xenopus laevis. Nature 219, 454–463 (1968).
Grunstein, M. & Hogness, D.S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl Acad. Sci. USA 72, 3961– 3965 (1975).
Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503– 517 (1975).
Kafatos, F.C., Jones, C.W. & Efstratiadis, A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 24, 1541–1552 (1979).
Lennon, G.G. & Lehrach, H. Hybridization analyses of arrayed cDNA libraries. Trends Genet. 7, 314– 317 (1991).
Khrapko, K.R. et al. An oligonucleotide hybridization approach to DNA sequencing. FEBS Lett. 256, 118–122 (1989).
Livshits, M.A. & Mirzabekov, A.D. Theoretical analysis of the kinetics of DNA hybridization with gel–immobilised oligonucleotides. Biophys. J. 71, 2795–2801 (1996).
Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P. & Trent, J. Expression profiling using cDNA microarrays. Nature Genet. 21, 10–14 (1999).
Chakravarti, A. Population genetics—making sense out of sequence. Nature Genet. 21, 56–60 ( 1999).
Milner, N., Mir, K.U. & Southern, E.M. Selecting effective antisense reagents on combinatorial. Nature Biotechnol. 15, 537– 541 (1997).
Lipshutz, R.J., Fodor, S.P.A., Gingeras, T.R. & Lockhart, D.J. High density synthetic oligonucleotide arrays. Nature Genet. 21, 20–24 (1999).
Blanchard, A.P., Kaiser, R.J. & Hood, L.E. Synthetic DNA arrays. Biosensors and Bioelectronics 11, 687–690 ( 1996).
Maskos, U. & Southern, E.M. A novel method for the analysis of multiple sequence variants by hybridisation to oligonucleotide arrays. Nucleic Acid Res. 21, 2267– 2268 (1993).
Southern, E.M., Maskos, U. & Elder, J.K. Analyzing and comparing nucleic acid sequences by hybridization toarrays of oligonucleotides: evaluation using experimental models. Genomics 13, 1008–1017 ( 1992).
Maskos, U. & Southern, E.M. A study of oligonucleotide reassociation using large arrays of oligonucleotides synthesized on a glass support. Nucleic Acids Res. 21, 4663–4669 (1993).
Southern, E.M. et al. Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res. 22, 1368–1373 (1994).
Gray, D.E., Case–Green, S.C., Fell, T.S., Dobson, P.J. & Southern, E.M. Ellipsometric and interferometric characterization of DNA probes immobilised on a combinatorial array. Langmuir 13, 2833–2842 ( 1997).
Guo, Z., Guilfoyle, R.A., Thiel, A.J., Wang, R. & Smith, L.M. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22, 5456– 5465 (1994).
Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P. & Trent, J. Expression profiling using cDNA microarrays. Nature Genet. 21, 10–14 (1999).
Cheung, V.G. et al. Making and reading microarrays. Nature Genet. 21, 15–19 (1999).
Maskos, U. & Southern, E.M. Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesized in situ. Nucleic Acids Res. 20, 1679–1684 (1992).
Matson, R.S., Rampal, J., Pentoney, S.L. Jr., Anderson, P.D. & Coassin, P. Biopolymer synthesis on polypropylene supports: oligonucleotide arrays. Anal. Biochem 224, 110–106 ( 1995).
Shchepinov, M.S., Case–Green, S.C. & Southern, E.M. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acid Res. 25, 1155–1161 (1997).
Williams, J.C., Case–Green, S.C., Mir, K.U. & Southern, E.M. Studies of oligonucleotide interactions by hybridisation to arrays: the influence of dangling ends on duplex yield. Nucleic Acids Res. 22, 1365–1367 (1994).
Maskos, U. & Southern, E.M. Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res. 20, 1675–1678 (1992).
Wood, W.I., Gitschier, J., Laskey, L.A. & Lawn, R.M. Base composition–independent hybridization in tetramethylammonium chloride: A method for oligonucleotide screening of highly complex gene libraries. Proc. Natl Acad. Sci. USA. 82, 1585– 1588 (1985).
Jacobs, K.A. et al. The thermal stability of oligonucleotide duplexes is sequence independent in tetraalkylammonium salt solutions: application to identifying recombinant DNA clones. Nucleic Acids Res. 16, 4637–4650 (1988).
Mir, K.U. Novel approaches for the analysis of nucleic acids. (D. Phil. thesis, Oxford University 1995).
Wetmur, J.G. & Davidson, N. Kinetics of renaturation of DNA. J. Mol. Biol. 31, 349– 370 (1968).
Nikiforov, T.T. et al. Genetic bit analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 22, 4167–4175 (1994).
Shchepinov, M.S., Udalova, I.A., Bridgman, A.J. & Southern, E.M. Oligonucleotide dendrimers: synthesis and use as polylabelled DNA probes. Nucleic Acid Res. 25, 4447– 4454 (1997).
Pastinen, T., Kurg, A., Metspalu, A., Peltonen, L. & Syvanen, A.C. Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 7, 606–614 (1997).
Nickerson, D.A., Kaiser, R., Lappin, S., Stewart, J. & Hood, L. Automated DNA diagnostics using an ELISA–based oligonucleotide ligation assay. Proc. Natl Acad. Sci. USA 87, 8923–8927 (1990).
Landegren, U., Kaiser, R., Sanders, J. & Hood, L. A ligase–mediated gene detection technique. Science 241, 1077 –1080 (1988).
Fotin, A.V., Drobyshev, A.L., Proudnikov, D.Y., Perov, A.N. & Mirzabekov, A.D. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 26, 1515–1521 ( 1998).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Southern, E., Mir, K. & Shchepinov, M. Molecular interactions on microarrays. Nat Genet 21 (Suppl 1), 5–9 (1999). https://doi.org/10.1038/4429
Issue Date:
DOI: https://doi.org/10.1038/4429
This article is cited by
-
Genome-wide Identification of DNA-protein Interaction to Reconstruct Bacterial Transcription Regulatory Network
Biotechnology and Bioprocess Engineering (2020)
-
Target capturing performance of microfluidic channel surface immobilized aptamers: the effects of spacer lengths
Biomedical Microdevices (2019)
-
Comparison of mitochondrial gene expression and polysome loading in different tobacco tissues
Plant Methods (2017)
-
Hybridization of poly(rI) with poly(rC) adsorbed to the carbon nanotube surface
Nanoscale Research Letters (2014)