Article Text
Abstract
Background Multiple morphological abnormalities of the flagella (MMAF) consistently lead to male infertility due to a reduced or absent sperm motility defined as asthenozoospermia. Despite numerous genes recently described to be recurrently associated with MMAF, more than half of the cases analysed remain unresolved, suggesting that many yet uncharacterised gene defects account for this phenotype
Methods Exome sequencing was performed on 167 infertile men with an MMAF phenotype. Immunostaining and transmission electron microscopy (TEM) in sperm cells from affected individuals were performed to characterise the ultrastructural sperm defects. Gene inactivation using RNA interference (RNAi) was subsequently performed in Trypanosoma.
Results We identified six unrelated affected patients carrying a homozygous deleterious variants in MAATS1, a gene encoding CFAP91, a calmodulin-associated and spoke-associated complex (CSC) protein. TEM and immunostaining experiments in sperm cells showed severe central pair complex (CPC) and radial spokes defects. Moreover, we confirmed that the WDR66 protein is a physical and functional partner of CFAP91 into the CSC. Study of Trypanosoma MAATS1’s orthologue (TbCFAP91) highlighted high sequence and structural analogies with the human protein and confirmed the axonemal localisation of the protein. Knockdown of TbCFAP91 using RNAi impaired flagellar movement led to CPC defects in Trypanosoma as observed in humans.
Conclusions We showed that CFAP91 is essential for normal sperm flagellum structure and function in human and Trypanosoma and that biallelic variants in this gene lead to severe flagellum malformations resulting in astheno-teratozoospermia and primary male infertility.
- reproductive medicine
- genetics
- molecular genetics
Statistics from Altmetric.com
Introduction
Spermatozoa are highly differentiated haploid cells resulting from spermatogenesis, a complex developmental process in which germline stem cells undergo a series of divisions and morphological modifications. At the simplest level, mature spermatozoa are polarised cells composed of a head and a flagellum joined by the connecting piece. Each of these parts contains highly specialised structural features reflecting the unique functions required for fertilisation. The flagellum contains the energy sources and machinery to generate the motility necessary for the spermatozoa to reach the egg. Flagellum integrity is thus critical for normal sperm function and sperm flagellum defects consistently lead to male infertility due to a reduced or absent sperm motility defined as asthenozoospermia. Asthenozoospermia is a frequent sperm phenotype found in approximately 19% of infertile men1 and can be combined with morphological sperm defects. Since 1984, morphological abnormalities of the sperm flagella leading to asthenozoospermia have been regularly reported. This rare phenotype has been subsequently reported as ‘dysplasia of the fibrous sheath’, ‘short tails’ or ‘stump tails’. In 2014, this heterogeneous group of flagellar defects was called ‘MMAF’ for multiple morphological anomalies of the flagella to standardise all these terms.2 Exome sequencing allowed us to partially elucidate genetic and physiopathological mechanisms leading to sperm flagellum defects. To date, variants in at least 16 genes have been found to be associated with MMAF phenotype.3–17 We however observe that despite regular new gene identification, about two-thirds of MMAF individuals remain unresolved demonstrating the high genetic heterogeneity of this phenotype.4 Several other genes are therefore likely to be involved in this phenotype and further efforts are needed to improve our genetic understanding of the molecular pathogeny of asthenozoospermia.
Materials and methods
Patients
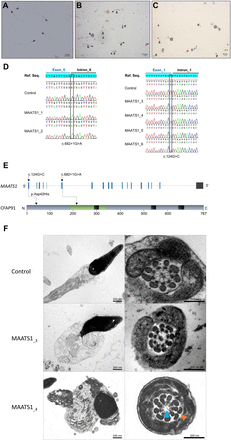
With the aim of identifying further genetic causes associated with human asthenozoospermia due to MMAF, we analysed whole exome sequencing data from a cohort of 167 patients with MMAF previously established by our team.4 All patients presented with a typical MMAF phenotype characterised by severe asthenozoospermia (total sperm motility below 10%) with at least three of the following flagellar abnormalities present in >5% of the spermatozoa: short, absent, coiled, bent or irregular flagella.4 All patients had a normal somatic karyotype (46,XY) with normal bilateral testicular size, normal hormone levels (FSH, testosterone and prolactin) and secondary sexual characteristics. All these patients presented only with isolated infertility without any other clinical features. Sperm analysis was carried out in the source laboratories during routine biological examination of the patients according to WHO guidelines.18 The morphology of the patients’ sperm was assessed with Papanicolaou staining (figure 1A–C). Detailed semen parameters of the six MAATS1 affected individuals are presented in table 1. Sperm samples for additional phenotypic characterisation could only be obtained from MAATS1_3 and MAATS1_4. The six patients were (MAATS1_1 to MAATS1_6) were initially recruited at the Clinique des Jasmin in Tunis (Tunisia) and were originated from North Africa. All these patients were born to first-cousin parents.
Morphology of normal and MAATS1 mutant spermatozoa and the variants identified in MAATS1-mutant patients. (A–C) Light microscopy analysis of spermatozoa from fertile control individuals (A), patient MAATS1_3 (B) and patient MAATS1_4 (C). Most spermatozoa from MAATS1 patients have flagella that are short (#), absent (*), coiled (@) or of irregular calibre (&). Head malformations were also observed (δ). (D) Electropherograms from Sanger sequencing indicating the homozygous state of the two identified variants: the c.682+1G>A variant (NM_033364.3) affecting the splice donor site following exon 6 and the c.124G>C; p.(Asp42His) variant (NM_033364.3) affecting the last nucleotide of exon 1 in patients MAATS1_1-2 and MAATS1_3-6, respectively. Variants are annotated following the Human Genome Variation Society (HGVS) recommendations. (E) Structure of the canonical transcript of MAATS1 showing the position of the observed variants. The functional structure of the encoded protein is shown in the lower panel. Coiled-coil domains are indicated by black boxes. CFAP91-domain (IPR032840) is highlighted in green. (F) Transmission electron microscopy analyses of sperm cells from a control individual and the two patients MAATS1_3 and MAATS1_4. In longitudinal sections, we can notice a short tail corresponding to a cytoplasmic mass containing the different components of the flagellum, all disorganised. In cross-sections of sperm flagellum from MAATS1 patients, we can observe some sections showing a 9+0 axoneme lacking the CPC (blue arrow) with an abnormal number of outer dense fibres (orange arrow) (MAATS1_4) as well as some sections with a normal axonemal 9+2 conformation (MAATS1_3). Scale bars: 200 nm.
Detailed semen parameters for the six individuals with MMAF harbouring homozygous MAATS1 variant
Exome sequencing and bioinformatic analysis
Data processing of the whole cohort of 167 patients with MMAF were performed according to our previously described protocol.4 For details, see online supplementary methods.
Supplemental material
Sanger sequencing
MAATS1 variants identified by exome sequencing were validated by Sanger sequencing as previously described.4 PCR primers and protocols used for each patient are listed in the online supplementary table S1.
Real-time RT-qPCR analysis
RT-qPCR was performed with cDNAs from a panel of 10 human tissues purchased from Life Technologies as previously described.4 RT-qPCR was performed as described in the online supplementary methods. The primer sequences are presented in the online supplementary table S2.
Transmission electron microscopy analysis of human sperm cells
Transmission electron microscopy (TEM) experiments were performed using sperm cells from control individuals and from MAATS1_3 and MAATS_4, both carrying the missense variant c.124G>C; p.(Asp42His). TEM was performed as described in the online supplementary methods.
Minigene assays
All recombinant plasmid constructs were produced as described previously19 and details about MAATS1 minigene assay are presented in the online supplementary methods. Briefly, the wild-type MAATS1 plasmid construct (pSplicePOLR2G–MAATS1_WT) was generated by cloning sequence of exon 1, 5’ untranslated region and the intronic flanking regions of MAATS1 into the pSplicePOLR2G vector. A c.124G>C variant was also introduced in the pSplicePOLR2G–MAATS1_ c.124G>C vector by using the QuickChange Site-Directed mutagenesis kit according to the manufacturer’s instructions (Agilent Technologies, Santa Clara, California, USA). Sequencing analyses were performed to check the integrity of all of the plasmid constructs.
Immunostaining in human sperm cells
Immunofluorescence (IF) experiments were performed using sperm cells from control individuals and from MAATS1_3 and MAATS_4, both carrying the missense variant c.124G>C; p.(Asp42His). For each studied patient with MMAF, 200 sperm cells were analysed by two different experienced operators and the IF staining intensity and pattern were compared with a fertile control. IF experiments were performed as described in the online supplementary methods. The list of primary antibodies used in the present study is detailed in the online supplementary table S3.
Trypanosoma brucei culture and transfection
The trypanosome cell lines used in this study derived from the procyclic form parental Trypanosoma brucei SmOxP427 strain co-expressing the T7 RNA polymerase and the tetracycline repressor and were grown and transfected as described.17 When required, the culture medium was supplemented with puromycin (1 µg/mL), hygromycin (25 µg/mL), phleomycin (5 µg/mL). RNA interference (RNAi) was induced with tetracycline (10 µg/mL).
T. brucei cell lines
We generated in SmOxP427 cells a TbCFAP91 transgene at its endogenous locus encoding N-terminal 10 HA epitope-tagged TbCFAP91 (HA::TbCFAP91) using a pPOTv7-blast-10HA vector as described.17 For TbCFAP91 RNAi, bp 1–864 were cloned between the XhoI and XbaI sites of p2T7tiB and transfected into the HA::TbCFAP91-expressing background cell line. Primers are described in the online supplementary table S4.
Immunofluorescence of Trypanosoma cells
Cells were collected, washed and processed for immunolabelling on methanol fixed detergent-extracted cells (cytoskeleton (CSK)) as described by Albisetti et al. The antibodies used are described in the online supplementary table S5. Images were acquired on a Zeiss Imager Z1 microscope, using a Photometrics Coolsnap HQ2 camera, with a 100x Zeiss objective (NA 1.4) using Metamorph software (Molecular Devices), and processed with ImageJ.
Sedimentation assays and video-microscopy
Sedimentation assays and videos were done as described.20 Briefly, the cells were placed in cuvettes and incubated 24 hours without shaking. The optical density (OD600 nm) was measured before mixing (ODb, the cell density reflects the ‘swimming’ cells) and after mixing (ODa, the cell density reflects ‘swimming’ and ‘sedimenting’ cells). The graphs presented in figure 1F represent the percentage of swimming cells calculated as 100−(ODb/ODa)×100 and, for the RNAi cells, normalised with the HA::TbCFAP91 parental cell line. Video-microscopy was carried out on a Zeiss AxioImager Z1, 40x lens (NA 1.3). 25 s of digital video from separate regions were captured and analysed using Metamorph software (Molecular Devices) (speed x4).
Electron microscopy of Trypanosoma cells
Electron microscopy blocks of cells and thin sections were prepared as described.21 Thin sections were visualised on a FEI Tecnai 12 electron microscope, camera ORIUS 1000 11M Pixel (resolution 3±5 nm). Images were acquired with DigitalMicrograph and processed with ImageJ.
Western blot analysis
Proteins from whole cell extracts or detergent-extracted cells and flagella were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and semi-dry transferred and further processed as described.21 The antibody used and dilutions are described in the online supplementary table S5.
Results
Exome sequencing identified homozygous variants in MAATS1 in patients with MMAF
In the whole cohort of 167 patients with MMAF, we previously identified 60 patients (36 %) with harmful variants in known MMAF-related genes.4 7 9 10 13 15–17 22 After reanalysis of the remaining exomes, we identified six additional unrelated patients originated from North Africa (MAATS1_1-6) with a homozygous variant in MAATS1 (MIM: 609910), a gene not previously associated with any pathology, accounting for 3.6% of our cohort. Two patients (MAATS1_1,2) had a splice-site variant c.682+1G>A (NM_033364.3, GRCh38 g.119715744G>A, rs147597066) and four (MAATS1_3-6) had a missense variant c.124G>C; p.(Asp42His) (NM_033364.3, GRCh38 g.119703222G>C, rs149348782) located in a splice-site region (figure 1, table 1 and online supplementary table S6). All variants identified by exome sequencing were validated by Sanger sequencing as illustrated in figure 1D. All MAATS1 variants are deposited in ClinVar under reference SUB6200231. No other candidate variants reported to be associated with cilia, flagella or male fertility were present in any of the six MAATS1 affected individuals. No heterozygous MAATS1 variants were found in our full cohort.
MAATS1 (also known as CFAP91, AAT-1 or c3orf15) is located on chromosome 3 and contains 18 exons encoding cilia-associated and flagella-associated protein 91 (CFAP91), a predicted 767-amino acid protein (Q7Z4T9). MAATS1 is mostly expressed in the pituitary and the testis according to data from GTEx and described to be associated with cilia and flagella.23 Moreover, quantitative single-cell RNA sequencing datasets from human adult testis (ReproGenomics Viewer)24 indicate an abundant expression in the germ cells from zygotene spermatocyte to late spermatid stages suggesting a role in sperm cell differentiation and/or function. In addition, the encoded CFAP91 protein was significantly detected in human sperm proteome,25 whereas it was found at a very low level in human airway cilia.26 RT-qPCR experiments performed with a panel of 10 human tissues including other ciliated tissues such as trachea confirmed these results, showing that MAATS1 transcripts are largely overexpressed in the testis compared with all the other tested tissues (online supplementary figure S1).
The first variant c.682+1G>A identified in patients MAATS1_1 and MAATS1_2 is a splice variant which changes the invariable +1 guanine and, thus, abolishes the intron 6 splice donor site according to the splice-site prediction algorithm Human Splicing Finder (http://www.umd.be/HSF3). The variant is present in the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/variant/3-119434591-G-A) with a minor allele frequency of 3.98×10−6 (online supplementary table S6). The second variant identified in four patient (MAATS1_3-6) is a missense variant c.124G>C; p.(Asp42His) located in exon 1 (figure 1E). This variant is present at a very low frequency in the general population (8.42×10-5 in gnomAD database/3–119422069 G-C) (online supplementary table S6) and is absent from our in-house database of 94 control exomes obtained from other infertile patients from North Africa (n=60) and the Middle East (n=34) (corresponding to the geographical origin of most patients in this study) presenting with a different sperm phenotype (azoospermia). Using prediction software for non-synonymous SNPs, we found that this missense change is predicted to be deleterious by the Sorting Intolerant from Tolerant (SIFT) algorithm (score of 0) and probably damaging by PolyPhen (score of 0.999) (online supplementary table S6). Moreover, the concerned amino acid (Asp42) was found conserved in CFAP91 orthologues (online supplementary figure S2). The in silico splicing prediction module in the Alamut software V.2.10 (Interactive Biosoftware) was used to predict the effect of the variant c.124G>C on the effectiveness of splicing. This software regroups five splice-site prediction tools: SpliceSiteFinder-like; MaxEntScan; NNSPLICE; GeneSplicer and Human Splicing Finder. Interestingly, the c.124G>C variant which concerns the last nucleotide of MAATS1 exon 1 was predicted to weaken the consensus splice donor site of MAATS1 exon 1 (online supplementary figure S3). Another tool for predicting variant spliceogenicity was also used: SPiCE,27 accessible at https://sourceforge.net/projects/spicev2-1/. SPiCE uses logistic regression by running different combinations of in silico tools and reveals an unprecedented sensitivity and specificity of 99.5% and 95.2%, respectively. The variant was predicted to alter splicing with a SPiCE probability of 0.99196 (online supplementary table S6).
In vitro splicing analysis of the missense variant c.124G>C; p.(Asp42His)
In order to confirm the bioinformatic predictions and because mRNA from affected individuals was not available, we tested the consequences of the c.124G>C; p.(Asp42His) variant using a splicing minigene assay. HEK293T cells were transiently transfected with different pSplicePOLR2G–MAATS1 minigenes (online supplementary figure S4A), and then the splicing pattern of the mutant minigenes was compared with that of the wild type. Transfection of the pSplicePOLR2G–MAATS1_WT minigene in HEK293 cells resulted in the production of one major mRNA product (online supplementary figure S4B). Sequencing analysis confirmed that this product consisted of the succession of exon 1 of MAATS1 and exon 2 of PolR2G (online supplementary figure S4C). RT-PCR analysis of the c.124G>C variant in exon 1 (pSplicePOLR2G–MAATS1_ c.124G>C) resulted in the synthesis of a product that differs in size from those generated with the wild-type construct (online supplementary figure S4B). Direct sequencing of the band indicated a 49 bp partial retention of intron in mRNA: r.[124G>C; 124_125ins124+1_124+49] (online supplementary figure S4B). A second plasmid construct was generated for the study with the first three exons of MAATS1 as well as the 5’ and 3’ intronic flanking regions and confirmed these results (data not shown). Overall, these data indicate that the c.124G>C variant alters the cellular splicing of a full-length mature transcript, and leads to an aberrant transcript with a 49 bp insertion inducing a stop codon after the first exon. These observations permit to define at the protein level a new nomenclature for this genetic variant: p.Asp42ArgfsTer15.
MAATS1 variants lead to a severe axonemal disorganisation of the sperm flagellum
Using TEM, we first studied the ultrastructure of sperm cells from the affected individuals MAATS1_3 and MAATS_4 both carrying the missense variant c.124G>C; p.(Asp42His) (figure 1F). Due to the low number of available sperm cells, only a few cross-sections (<10) had a sufficient quality to be analysed and therefore no statistical analysis could be done. Among these cross-sections, we observed various defects from normal sections (figure 1F) to abnormal conformations of the axoneme with in particular the absence of the central pair complex (CPC) (9+0 conformation) (figure 1F). Observation of longitudinal sections showed severe flagellum abnormalities, which appeared completely disorganised, resulting in truncated flagella or a cytoplasmic mass encompassing unassembled axonemal components (figure 1F). In some sections, we also observed peri-axonemal structural defects such as an abnormal number of outer dense fibres (figure 1F), a defect already observed in sperm from MMAF-affected patients carrying variants in other genes.
To define the ultrastructural defects established by TEM and to characterise the molecular defects induced by the c.124G>C MAATS1 variant in human sperm, we subsequently studied the presence and localisation of several proteins belonging to different axonemal substructures by IF. The presence of the following proteins was investigated: SPAG6 as a marker of the CPC, DNAI2 and DNALI1 as markers outer dynein arm (ODA) and inner dynein arm (IDA), respectively, RSPH1 as a marker of the radial spokes (RS). We observed that in sperm from patients MAATS1_3 and MAATS1_4, staining of SPAG6 was heterogeneous: the staining was present in most sperm with a long flagellum but it was totally absent in sperm cells with short tails (figure 2). When SPAG6 staining was observed, the signal along the flagellum appeared thinner and more irregular than in the control (figure 2A). This finding suggests that the CPC is inconsistently present in sperm cells from the affected individuals as established by TEM. Moreover, we observed that the presence of CPC seems to be directly related to the sperm flagellum morphology. The staining of RSPH1 was totally absent in sperm with short flagella or dramatically reduced in sperm cells with long flagella (figure 3A,B); in the later cases, RSPH1 staining displayed an abnormal dotted and irregular pattern. In contrast, immunostaining for DNAI2 and DNALI1 were similar to controls irrespective of the sperm morphology (short or long flagella), indicating that ODAs and IDAs were not directly affected by variants in MAATS1 (online supplementary figures S5 and S6).
The central pair complex (CPC) is impacted in MAATS1-mutant patients. (A) Sperm cells from a fertile control individual and from MAATS1_3 and MAATS1_4 patients stained with anti-SPAG6 (HPA038440, Sigma-Aldrich, rabbit, 1:500, green), a protein located in the CPC and anti-acetylated tubulin (32–2500, ThermoFisher, mouse, 1:1000, red) antibodies. DNA was counterstained with 4',6-diamidino-2-phenylindole (DAPI II). The SPAG6 immunostaining is uniformly present throughout the flagellum in control sperm cells. Contrary to the control, sperm cells from MAATS1 patients exhibit a heterogeneous staining depending on flagellar morphology. Scale bars: 10 µm. (B) Histogram showing the percentage of analysed spermatozoa with or without SPAG6 staining according to their morphology (number of sperm cells analysed=200). Scale bars: 10 µm.
Radial spokes are affected in MAATS1-mutant patients. (A) Sperm cells from a fertile control individual and MAATS1_3 and MAATS1_4 patients stained with anti-RSPH1 (HPA017382, Sigma-Aldrich, rabbit, 1:100, green), a protein located at the head of the radial spokes and anti-acetylated tubulin (32–2500, ThermoFisher, mouse, 1: 1000, red) antibodies. DNA was counterstained with DAPI II. RSPH1 immunostaining is present throughout the flagellum in control sperm cells but is mainly absent or strongly reduced in MAATS1 patients. In a few sperm cells, the RSPH1 staining is present but displays an abnormal pattern with a dotted and irregular signal. Scale bars: 10 µm. (B) Histogram showing the percentage of analysed spermatozoa with or without RSPH1 staining according to their morphology (number of sperm cells analysed=200). Scale bars: 10 µm.
WDR66 is absent in sperm cells from patients with a pathogenic variant in MAATS1
In addition, we also examined the localisation of WDR66 (CFAP251), a protein localised in the calmodulin-associated and spoke-associated complex (CSC) at the base of radial spoke 3 in Tetrahymena and Chlamydomonas and described as a partner of CFAP91.28 In control sperm, WDR66 immunostaining decorated the full-length flagella, but in patients MAATS1_3 and MAATS1_4, WDR66 staining was weak or completely absent whereas tubulin staining remained detectable (online supplementary figure S7). This result demonstrates that the CSC is strongly disorganised, and is in agreement with the fact that CFAP91 and WDR66 (CFAP251) are two interacting proteins, likely to be closely associated within the same axonemal complex.
T. brucei CFAP91 is an axoneme-associated protein involved in flagellum motility and central pair positioning
To validate our candidate gene and to better characterise CFAP91 localisation and function, we decided to use a Trypanosoma model, which could be used for forward and reverse genetics and which has largely contributed to the characterisation of the molecular pathogeny of different human diseases caused by defective cilia and/or flagella.29 The T. brucei axoneme is similar to that of mammalian flagella with a 9+2 structure. T. brucei flagellum contains a para-axonemal structure named paraflagellar rod (PFR), which plays a role in flagellum motility and serves as a platform for metabolic and signalling enzymes, similar to para-axonemal structures FS and ODFs on mammalian sperm. Using BlastP analysis on the T. brucei genome database, we identified the putative T. brucei CFAP91 orthologue Tb927.11.2790 (named TbCFAP91 in this study). TbCFAP91 is a 709 amino acid protein annotated as a tetratricopeptide repeat putative protein. The calculated identity and similarity between human CFAP91 and TbCFAP91 is 27.97% and 46%, respectively. We thus generated a T. brucei cell line expressing N-terminal-HA tagged TbCFAP91 (HA::TbCFAP91) and localised the protein by IF on detergent-extracted cells (CSK). HA::TbCFAP91 localised specifically to the flagellum, as established by IF with the PFR protein, PFR2 (figure 4A) and by western blot analysis from flagellum fraction (figure 4B). HA::TbCFAP91 is associated with the axoneme fraction of the flagellum as its labelling is observed in flagellum part lacking PFR labelling. Furthermore, HA::TbCFAP91 labelling was observed at both the old flagellum and the new flagellum, suggesting that it is present at every stage of the cell cycle. To assess the functional role of TbCFAP91 in the parasite, we used a tetracycline-inducible RNAi system30 to knockdown the expression of TbCFAP91, and generated the HA::TbCFAP91/RNAi TbCFAP T. brucei cell line. Induction of RNAi by addition of tetracycline in the culture medium did not affect cell growth (figure 4C), but led to a decrease in HA::TbCFAP91 levels. Indeed, detection of HA::TbCFAP91 was strongly reduced by western blot analysis (figure 4D) and the protein was not detected by IF in the newly formed flagellum (figure 4E). Knockdown of TbCFAP91 did not affect the cell morphology and the flagella appeared normal in number, length and overall structure.31 However, induced cells were sedimenting, suggesting a flagellar beating defect (figure 4F). Indeed, sedimentation assays show that after 96 hours of induction, 90% of the induced cells sedimented. This sedimentation reflects a flagellar motility defect that was directly observed by video microscopy (online supplementary videos S1 and S2) and tracking (figure 4G). Remarkably, comparison of TEM images of thin sections of WT and 7 days RNAi-induced cells revealed that knockdown of TbCFAP91 induced a rotation of the central pair (CP)(figure 4H). Indeed, measurements of the α angle (see scheme in figure 4I) demonstrated that in WT cells, the CP orientation was fixed as previously shown,32 while in TbCFAP91 RNAi-induced cells some cross-sections exhibit a rotated CP axis (figure 4H,I).
Supplementary video
Supplementary video
TbCFAP91 is an axoneme-associated protein involved in flagellum motility and central pair positioning in Trypansoma brucei. (A) A late G2 cell expressing HA::TbCFAP91 was detergent-extracted and immunolabelled with anti-PFR2 (magenta) that labels the para-axonemal paraflagellar rod (PFR) structure and anti-HA that labels HA::TbCFAP91 (yellow). Both PFR2 and HA::TbCFAP91 are present at the old flagellum (OF) and the new flagellum (NF). The cell body is outlined by a dashed line. (B) Western blot analysis of HA::TbCFAP91 on whole cells (WC), detergent-extracted cells (cytoskeleton (CSK)) and salt-extracted cytoskeletons (flagella, FG) using anti-HA. Anti-TbSAXO was used as flagellum-specific protein control, and anti-enolase as cytoplasmic content control. (C) Comparative growth curve of parental (wild-type (WT)), non-induced and induced for RNA interference (RNAi) of TbCFAP91. (D) Immunolabelling of PFR (magenta) and HA::TbCFAP91 (yellow) on a detergent-extracted cell induced 24 hours for TbCFAP91 RNAi. The OF is still HA::TbCFAP91 positive while the NF is negative for HA::TbCFAP91 labelling, showing the efficiency and specificity of the RNAi. (E) Western blot analysis showing a decrease of HA::TbCFAP91 level after RNAi induction (non-induced (NI), induced (I)). (F) Percentage of sedimented cells in WT, NI and RNAi-I for 24–96 hours. Percentages of sedimentation of RNAi-induced cells were normalised to the HA::TbCFAP91 cell line. Data are represented as the mean±SEM. (G) Mobility tracking from online supplementary videos S1 and S2. The positions of patient cells are plotted at 0.2 s intervals. Arrowhead: ending position. Scale bars: 50 µm. (H) Electron micrograph of thin sections of whole WT cells and 7 days RNAi-induced cells. (I) RNAi of TbCFAP91 affects CP orientation. The angle α was measured in sections of WT cells (n=23) and sedimented RNAi-induced cells (n=43). The graph indicates the mean±SD. In A and E, the mitochondrial genomes and the nuclei are stained with DAPI (grey). Scale bars represent 5 µm in the main immunofluorescence figures, 1 µm in zoom insets and 200 nm in electron microscope.
Discussion
In the present work, we showed that the presence of biallelic MAATS1 variants induce a typical MMAF phenotype in humans, demonstrating that this gene is necessary for sperm flagellum structure. Several ultrastructural components of the axoneme were found to be affected by the absence of the encoded protein. First, CPC anomalies were shown by IF or TEM experiments (figures 1F and 2). We observed a heterogeneous pattern of CPC staining, and the presence of the CPC seems closely associated with the flagellar morphology, with a higher proportion of CPC defects arising in shorter flagellum (figure 2B). This observation supports the hypothesis that the CPC is a key component of the sperm flagellum ensuring a stable construction and/or a global cohesion of the full axonemal structure.2 The RS proteins seemed more severely and constantly affected by the MAATS1 variants even though few sperm cells showed normal RSPH1 staining (figure 3). RS enhance the stability of the CP and of the entire axoneme.33 When the RS are defective the CPC can be assembled but tends to be misplaced with CP transposition, shift or rotation.34 In Trypanosoma, we confirmed that CFAP91 is an axonemal protein and by RNAi experiments we observed that its absence leads to structural defects (figure 4). We indeed showed using TEM that flagella from RNAi-induced parasites exhibit an abnormal axoneme with a rotated CPC, a defect not observed in control trypanosome cell lines (figure 4H,I).
All these results are consistent with the known localisation of CFAP91 in the axoneme: CFAP91 is an A-kinase anchoring protein (AKAP)-binding protein and is a major component of the CSC.23 The CSC is located within the region that interconnects the bases of RS2, RS3 and the N-DRC.28 It mediates regulatory signals between the radial spokes and the dynein arms and therefore is involved in the regulation of flagellar motility.28 Based on the localisation of the CSC at the base of the radial spokes, it was expected that the absence of members of this complex would result in radial spoke assembly defects.23 In Chlamydomonas, knockdown of FAP91 by microRNAs results in ultrastructural defects in RS2 and RS3 and in a partial destabilisation of the N-DRC.35 Interestingly, knockout of each CSC subunit in Chlamydomonas or Tetrahymena led to a reduced RS density but not to the total absence of the RS indicating heterogeneity among the repeats.28 35 Such heterogeneity was also observed in sperm cells from MAATS1 patients, as illustrated by the RSPH1 staining (figure 3).
In Chlamydomonas, in addition to FAP91, the CSC consists of CaM and two other conserved proteins: FAP61 (also known as c20orf26) and FAP251 (known as WDR66).28 The two subunits FAP61 and FAP251 are within the base of the RS3 while the FAP91 subunit contacts the base of RS2 on the side facing RS3.28 We and another team previously demonstrated that deleterious variants in CFAP251/WDR66 (MIM: 618146) lead to a MMAF phenotype with similar flagellar defects to what is observed in MAATS1 affected individuals.17 36 We showed in the present work that CFAP251/WDR66) was totally absent from sperm cells from patients MAATS1_3 and MAATS1_4, supporting the close physical and functional interaction of these two proteins within the CSC (online supplementary figure S7). Last, CFAP91 interacts with the radial spoke protein 3 (RSP3), which is a conserved AKAP23 located in the spoke stalk/base of RS1 and RS2 and involved in the anchoring of the RS complex to the microtubule and to the CSC.37 Moreover, Chlamydomonas mutants lacking RSP3 (pf14) completely failed to assemble the radial spokes.38 Overall, these data suggest that CFAP91 is critical for radial spokes assembly and stability through its essential role within the CSC and its interaction with other axonemal partners such as WDR66 or RSP3. In humans, the absence of CFAP91 in sperm cells likely destabilises the RS structure and as a consequence the stability of the CPC, both of these defects resulting in a severe axonemal disorganisation responsible for a MMAF sperm phenotype.
In our cohort of 167 patients, we had identified 60 patients (36%) with variant in known MMAF-related genes. The diagnostic yield rises to 40% when adding the six new MAATS1-affected individuals, highlighting the performance of the exome sequencing strategy in the genetic exploration of patients with MMAF. MAATS1 is one of the most frequently mutated genes in our cohort with 3.6% of patients. Combined with patients with a WDR66 alteration (11 patients), variants in CSC-related genes are found in 10.2% of our cohort, suggesting that the CSC is the main structure affected in patients with MMAF. T. brucei was chosen to validate our candidate gene because this flagellated protozoan has an axonemal organisation similar to human with a 9+2 structure.39 Moreover, the T. brucei flagellum also contains the para-axonemal PFR structure, which plays a role in flagellum motility and serves as a platform for metabolic and signalling enzymes, similar to the mammalian sperm peri-axonemal structures, the fibrous sheath and outer dense fibres.40 Overall, these results demonstrated that the combined use of exome sequencing and Trypanosoma model is an efficient strategy to identify new candidate gene responsible for MMAF phenotype in human.
Altogether, these data demonstrate that CFAP91 is essential for normal sperm flagellum structure and function in human and the flagellated protist T. brucei, and that variants in MAATS1 lead to severe flagellum malformations resulting in astheno-teratozoospermia and primary male infertility. Interestingly, a mouse model inactivated for the Maats1 gene is also available from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/allele/MGI:6152510). Infertility is reported in male mice but no additional details about the sperm phenotype are currently provided.
Acknowledgments
The authors would like to thank all individuals for their participation. The authors would like to thank Samuel Dean (Oxford University) for the pPOT vector, Nicolas Biteau (Bordeaux University) for the anti-PFR2 antibody and Frédéric Bringaud (Bordeaux University) for the anti-enolase antibody.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
GM and JB contributed equally.
PR and CCo contributed equally.
Contributors All authors of this manuscript fulfil the criteria of authorship. VS, ED and RZ recruited patients and collected clinical information. CCa, MB, Z-EK, GLG, CK, IG, YF and GP performed molecular analyses. GM and JB performed IF experiments. NT-M performed the bioinformatic analysis. DD, DRR and MB performed the Trypanosoma study. AT and ED performed the TEM experiments. GM, JB, AT, CA, MB, PFR and CCo performed data analysis and interpretation. GM, JB, AT, CA, MB, PFR and CCo designed the study and wrote the manuscript.
Funding This work was mainly supported by the following grants by the Agence Nationale de la Recherche (ANR) (MUCOFERTIL 12-BSV1–0011 and MASFLAGELLA 14-CE15), the LabEx ParaFrap (ANR-11-LABX-0024), the 'Whole genome sequencing of subjects with Flagellar Growth Defects (FGD)' financed by the fondation maladies rares (FMR) for the programme Séquençage à haut débit 2012. The electron microscopy analysis was done in the Bordeaux Imaging Centre, a service unit of the CNRS/INSERM and Bordeaux University, member of the national infrastructure France BioImaging supported by the ANR (ANR-10-INSB-04).
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available on reasonable request.