Article Text
Abstract
Mutation of BRCA1 and BRCA2 is the most common cause of inherited breast and ovarian cancer. Genetic screens to detect carriers of variants can aid in cancer prevention by identifying individuals with a greater cancer risk and can potentially be used to predict the responsiveness of tumours to therapy. Frequently, classification cannot be performed based on traditional approaches such as segregation analyses, including for many missense variants, which are therefore referred to as variants of uncertain significance (VUS). Functional assays provide an important alternative for classification of BRCA1 and BRCA2 VUS. As reviewed here, both of these tumour suppressors promote the maintenance of genome stability via homologous recombination. Thus, related assays may be particularly relevant to cancer risk. Progress in implementing functional assays to assess missense variants of BRCA1 and BRCA2 is considered here, along with current limitations and the path to more impactful assay systems. While functional assays have been developed to independently evaluate BRCA1 and BRCA2 VUS, high-throughput assays with sufficient sensitivity to characterise the large number of identified variants are lacking. Additionally, because of relatively low conservation of certain domains of BRCA1, and of BRCA2, between humans and rodents, heterologous expression in rodent cells may have limited reliability or capacity to assess variants present throughout either protein. Moving forward, it will be important to perform assays in human cell lines with relevance to particular tumour types, and to strengthen risk predictions based on multifactorial statistical analyses that also include available data on cosegregation and tumour pathology.
- hereditary breast and ovarian cancer
- BRCA1
- BRCA2
- variants of uncertain significance
- functional classification
Statistics from Altmetric.com
- hereditary breast and ovarian cancer
- BRCA1
- BRCA2
- variants of uncertain significance
- functional classification
BRCA1 and BRCA2 as cancer genes
In the 1990s, pathogenic variants in BRCA1 and BRCA2 were found to be associated with hereditary breast and ovarian cancer (HBOC).1 2 Among genes associated with HBOC, BRCA1 and BRCA2 confer the highest lifetime risks of these cancers and are the most frequently mutated genes in women with HBOC.3–6 Other cancers also show elevated incidence of mutations in BRCA1 (melanoma and testicular) and BRCA2 (male breast cancer, prostate cancer and pancreatic cancer).7
Clinical benefits of genetic testing for BRCA1 and BRCA2
Genetic testing for pathogenic variants in BRCA1, BRCA2 and other cancer susceptibility genes is recommended for individuals with a strong family and/or personal history of HBOC (see figure 1), since risk preventative strategies improve outcomes. Indeed, individuals who carry a pathogenic variant in BRCA1 and BRCA2 are recommended to consider prophylactic surgeries including bilateral risk-reducing mastectomy (RRM) and/or risk-reducing bilateral salpingo-oophorectomy (rrBSO). Individuals undergoing RRM are thought to reduce their risk of breast cancer by 85%–100%.8 Ovarian cancer risk is estimated to decrease by 69%–100% in BRCA1 carriers and BRCA2 carriers who undergo rrBSO.8 9 Importantly, screening for ovarian cancer is not as efficacious as for breast cancer. Therefore, since individuals with ovarian cancer are more likely to be diagnosed at later stages, rrBSOs are strongly suggested for BRCA1 and BRCA2 mutation carriers at age 35–40 or when childbearing is complete.10 Additionally, rrBSOs also lead to a decreased risk of breast cancer in women with pathogenic BRCA variants.8
Decision-making tree for when to sequence BRCA1/2 and for utilisation of the results. The decision-making tree for prevention and/or treatment recommendations is outlined for three scenarios in which clinical testing for BRCA1/2 is routinely done. HBOC, hereditary breast and ovarian cancer; PARP, poly(ADP-ribose) polymerase; VUS, variants of uncertain significance.
Knowledge of BRCA1 and BRCA2 pathogenic mutations also helps to guide cancer treatment. Tumours with loss of functional BRCA1 and/or BRCA2 are responsive to chemotherapeutic agents that induce DNA damage, such as cisplatin.11–14 More recently, the US Food and Drug Administration has approved the poly(ADP-ribose) polymerase (PARP) inhibitors, olaparib and rucaparib, for treating certain individuals with advanced ovarian cancer whose tumours have either germline or somatic mutations in BRCA1 or BRCA2.15–17 PARP inhibitors appear to be particularly efficacious in tumours that have suppression of DNA repair due to defects in the homologous recombination (HR) pathway, such as tumours lacking functional BRCA1 or BRCA2.18 Given the efficacy of PARP inhibitors, the current recommendations are that individuals with ovarian cancer have BRCA1/2 germline and/or tumour testing in order to direct therapy. As these recommendations are being incorporated into clinical care, the number of individuals having germline and/or somatic genetic testing for these genes is increasing.
Changes in the landscape of testing of BRCA1 and BRCA2
The landscape of genetic testing for BRCA1 and BRCA2, and other cancer susceptibility genes, is changing. For example, there are calls by some experts for doing population-based screening of the BRCA1 and BRCA2 genes of all women of a certain young age in order to prevent these cancers from occurring.19 Multiple factors underlie the current landscape of genetic testing. These include increased awareness of these genes by the public, changes in patent laws for BRCA1 and BRCA2, technological changes enabling the testing of multiple genes at cheaper costs, and testing of tumour DNA to identify either germline or somatic changes as a means to guide therapy. In particular, in June 2013, after hearing the case of the Association for Molecular Pathology v Myriad Genetics, Inc, the Supreme Court of the United States ruled that genes, which occur naturally, cannot be patented. This has led to multiple clinical molecular laboratories in the USA offering genetic testing for BRCA genes. Notably, competition in the market-place has helped make testing more affordable. Collectively, these events have led to increased rates of individuals being tested for mutations in BRCA1 and BRCA2,20 and an increased need for interpreting the significance of variants that are identified.
The major challenge associated with the vastly increased numbers of individuals who are having testing for BRCA1 and BRCA2 is that variants of uncertain significance (VUS) remain an issue. VUS, mainly missense alterations, are variants of unknown clinical impact; thus, individuals who carry a BRCA1/2 VUS do not have concrete information on which to make clinical decisions about cancer risk management or treatment. The effects of VUS on clinical decision-making, based on the results of genetic screens, are summarised in figure 1. This review will focus on DNA repair-related approaches to classifying variants, the pros and cons of the use of such functional assays for classification of BRCA1/2 variants and suggestions on how to move this field forward. In contrast to other reviews on the functional classification of BRCA1/2 variants,21 22 our up-to-date examination of the subject highlights the limited homology of rodent and human BRCA1 and BRCA2. As a result, we discuss the clear need to perform assays in human cells to enable the assessment of variants present anywhere within either protein.
Prevalence of germline and somatic VUS of BRCA1 and BRCA2
There are multiple databases including the Breast Information Core, ClinVar, Leiden Open Variation Database (LOVD) and the Global Alliance that catalogue germline genetic variants in BRCA1 and BRCA2 found through clinical sequencing. In particular, ClinVar is a database supported by the National Center for Biotechnology Information that annotates the clinical significance of human DNA sequence variants, as defined by the testing laboratory, locus-specific databases and/or an expert panel.23
Given that both BRCA1 and BRCA2 have key domains at or near their C-termini (figure 2A,B), nearly all nonsense and frameshift mutations result in truncations that can be classified as deleterious. The exception to this is variants in the last exon of the gene, which are not thought to result in nonsense-mediated decay, such as BRCA2 K3326X, and which are associated with only modest increases in cancer risk.24
A diagram of key structural domains of BRCA1 and BRCA2, all of which are involved in mediating DNA repair by homologous recombination, reflects the role of each protein. (A) Human BRCA1 has a RING domain from amino acids (a.a.) 9–98, a coiled-coil domain at a.a. 1393–1424 and two BRCT (BRCA1 C terminus) repeats (a.a. 1649–1734 and a.a. 1758–1859). Nuclear localisation signals (NLS) at a.a. 503–508 and 607–615 are not shown due to their small sizes. (B) BRCA2 contains a PALB2-binding domain (a.a. 10–40), and a RAD51-binding domain from a.a. 1008–2082 that includes 8 BRC repeats (grey bars) and intervening sequence (BRC1: a.a. 1008–1033, BRC2: 1218–1243, BRC3: 1427–1452, BRC4: 1523–1548, BRC5: 1670–1695, BRC6: 1843–1868, BRC7: 1977–2002 and BRC8: 2057–2082). BRCA2 also contains a DNA-binding domain (DBD) from a.a. 2402–3102, which is composed of a helical domain (a.a. 2402–2669) and three OB (oligonucleotide/oligosaccharide) folds (a.a. 2670–2803, 2809–3048 and 3056–3102), and a second RAD51-binding domain at the C-terminus (a.a. 3270–3305). Another DBD, which is less well defined than the C-terminal DBD, is present in BRCA2 within the region between a.a. 250 and 500. Two NLS at a.a. 3263–3269 and 3381–3385 are not shown. In both (A) and (B), domains that contain most of the variants of uncertain significance which have been functionally characterised utilising heterologous systems are shown in black or by patterned fill. These domains are also identified by a solid bar above the diagram. Most BRCA1 and BRCA2 VUS lie outside these better conserved domains.
There are currently ~5200 and ~6800 distinct germline variants of BRCA1 and BRCA2, respectively, listed in ClinVar. Of these, there are ~1450 BRCA1 missense mutations, of which~1300 are VUS and an additional 175 have conflicting interpretations. BRCA2, due in part to its larger size at 3418 amino acids, has ~2400 unique missense variants, of which ~2300 are VUS and ~320 have conflicting interpretations listed. Notably, for BRCA2, missense variants (~2400) are identified most frequently among alterations that affect the coding sequence. Frameshifts are observed with the next highest frequency (~1800), with nonsense (~550) and spice site (~140) variants being observed much less frequently. The list of VUS in BRCA1 and BRCA2 is likely an underestimate, as not all clinical testing laboratories, particularly those from outside of the USA, contribute data to ClinVar. Importantly, greater than 88% of BRCA1 missense variants and 95% of BRCA2 missense variants currently listed in ClinVar are unclassified, underscoring the abundant demand for approaches to classify them. Given how frequently they occur and the number that are currently uncharacterised, this review is largely focused on missense variants. It should also be noted that many of the missense variants of BRCA1 and BRCA2 that have been identified are not confined to recognised domains of either of the corresponding proteins (figures 3 and 4). Thus, strategies for classifying variants present anywhere within these proteins are greatly needed.
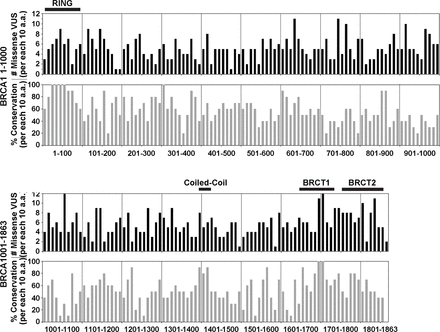
Missense VUS are distributed throughout BRCA1, including to regions of lower conservation. The number of distinct missense BRCA1 VUS listed in ClinVar is shown for groups of 10 amino acids (a.a.) (black bars, above) along with the % conservation between human and mouse BRCA1 for each group of 10 a.a. (grey bars, below). Vertical dividing lines indicate sets of 100 a.a. Key structural domains are shown by a horizontal black bar above plots of the frequency of missense VUS. Missense BRCA1 VUS that have been reported occur throughout the protein, regardless of the degree of conservation. The graph of percentage conservation demonstrates that there are areas of inexact conservation throughout the protein, including in the BRCT (BRCA1 C terminus) repeats, which have been tested more than most other domains using functional assays. Since inexact conservation could affect the reliability of assays that are based on heterologous expression of BRCA1 in rodent cells, this demonstrates potential advantages of expressing human BRCA1 in human cells. VUS, variants of uncertain significance.
Missense BRCA2 VUS are distributed throughout the protein, including to regions of lower conservation. (A) The number of distinct missense BRCA2 VUS listed in ClinVar is shown for groups of 10 amino acids (a.a.) (black bars, above) along with the % conservation between human and mouse BRCA2 for each group of 10 a.a. (grey bars, below). Vertical dividing lines indicate sets of 100 a.a. Key structural domains are shown by a horizontal bar above plots of the frequency of missense VUS. The BRC repeats are 25 a.a. motifs dispersed from a.a. 1008–2082 which bind RAD51. The C-terminal DNA-binding domain (DBD) is comprised of a helical domain and three OB (oligonucleotide/oligosaccharide) folds. Reported missense BRCA2 VUS are found throughout the protein regardless of the degree of conservation. The graph of percentage conservation demonstrates that there are areas of relatively low conservation throughout BRCA2, including part of the helical domain, which is in the C-terminal DBD that contains known pathogenic variants. Additionally, the BRC3, BRC5 and BRC6 motifs also have relatively low conservation. In contrast, there are other areas of high conservation that are not contained within known domains. Since inexact conservation could affect the reliability of assays based on heterologous expression of BRCA2 in rodent cells, this demonstrates a potential advantage of expressing human BRCA2 in human cells for assays. (B) Examples of pathogenic (R3052W) and likely pathogenic (L2647P) variants that do not display exact conservation in adjacent residues between human and mouse BRCA2. (C) Examples of benign variants lacking alignment or conservation with mouse BRCA2. Variant residues are indicated by asterisks, non-conserved residues in mouse are shown in grey, and residues without alignment in mouse BRCA2 are indicated by (-) in (B–C). VUS, variants of uncertain significance.
There is a demand for functional assays to classify BRCA1/2 variants
Since the inability to classify BRCA1 and BRCA2 variants limits the potential utility of screens of cancer genes to inform decisions concerning risk management, and given the prevalence of VUS of these genes, a means for better classification is greatly needed. The classification of missense variants in these genes is particularly difficult, as the genes are large and in silico approaches to classifying variants are not highly sensitive or specific.21 25 26 Traditionally, VUS in genes conferring Mendelian risk of disease have been classified as disease-associated (ie, pathogenic) using segregation analyses in families. This is a particularly powerful approach when a disease phenotype is relatively rare and multiple multigeneration families with several affected individuals carrying the same rare variant are available for study. Although some BRCA1/2 missense variants have been classified using this approach, it is not feasible for the vast majority of rare variants because:
many variants have only been reported in one or two families, thus there is not sufficient power to perform segregation analyses
known affected individuals in families are often deceased due to their cancer history
breast and ovarian cancer are relatively common in the general population, leading to phenocopies (individuals with the phenotype not due to the family mutation)
not all affected individuals in a family are interested in having genetic testing
not every individual with pathogenic variants gets cancer
some families have diagnoses of cancer on both maternal and paternal sides, making segregation difficult.
Another means that has been used successfully to classify hundreds of missense variants in BRCA1/2 as benign/neutral is allele frequency in large populations. The rationale for this strategy is that because unique pathogenic BRCA1 and BRCA2 missense changes are unlikely to be present at a frequency of greater than 0.1% or 1% in the population without phenotypic impacts, any variant in these genes that occurs in an unselected population at a high frequency is likely to be benign.
In silico prediction tools that incorporate features such as evolutionary conservation across species, how dramatic the predicted effect of amino acid substitution is on protein structure and whether the amino acid is positioned in a known functional domain have been used for characterisation of BRCA1/2 VUS. Many of these algorithms are not highly sensitive in isolation and tend to ‘over predict’ missense changes as pathogenic.27 Thus, there is a need to develop better in silico models that show higher specificity and sensitivity; it may be that models will need to be calibrated on a gene-by-gene basis.
To overcome difficulties in classifying BRCA1/2 VUS, the Evidence-Based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium was formed in 2009.26 This is an international collaborative group recognised as an expert panel by ClinVar for interpretation of BRCA1/2 variants. The strength of ENIGMA is that variants that may be seen only once or twice by a single group may be classified by pooling clinical and family segregation data across the entire consortium. Even within this large group, many variants are still too rare to classify based on family and clinical data. ENIGMA and others have developed multifactorial models that incorporate various data elements such as family history, cosegregation of disease in families, co-occurrence of a variant in trans with a previously classified pathogenic variant and histological features (figure 5).28–30 Over 100 BRCA1 and 100 BRCA2 variants have been classified using this approach.26 30
Overview of multifactorial model-based classification of missense VUS. Examples of types of input data used for multifactorial classification of BRCA1/2 missense VUS are described. The posterior probabilities recommended by IARC for the five classes of variant interpretations are noted. IARC, International Agency for Research on Cancer; VUS, variants of uncertain significance.
The International Agency for Research on Cancer Working Group (IARC), in collaboration with ENIGMA, developed a multifactorial five-tier classification scheme to classify BRCA1/2 VUS based on cosegregation with cancer in families, co-occurrence with previously identified pathogenic mutations and tumour histopathology, combined with an analysis of sequence conservation across species and the properties of mutated residues.31 IARC class 1 and class 2 designate non-pathogenic and likely non-pathogenic variants (probability ≤0.01 and >0.01 but ≤0.05), respectively. In contrast, classes 5 and 4 represent pathogenic and likely pathogenic variants (probability ≥0.99 and <0.99 but ≥0.95), respectively. Variants with intermediate probabilities (>0.05 but <0.95) remain unclassified and are designated as class 3, largely due to insufficient availability of family or other information (figure 5).
Despite the progress that has been made to develop systematic approaches to classifying missense variants, the vast majority of BRCA1/2 variants are VUS. The critical need for alternative approaches to classify VUS has led several research groups to develop in vitro assays that can be used to screen a variety of BRCA1 and BRCA2 mutations, including missense and in-frame deletions, for effects on BRCA1 and BRCA2 function.21 32–38 The beauty of functional assays is that they are not dependent on the existence of clinical or family data. Splice anomalies should be identified first and such variants excluded from subsequent DNA repair-related functional tests.
Domains and roles of BRCA1 and BRCA2 that can serve as the basis for functional assays
BRCA1 and BRCA2 are large proteins with functional domains distributed both N-terminally and C-terminally, as well as more centrally (figure 2). BRCA1 has a Really Interesting New Gene (RING) domain at its N-terminus (amino acids (a.a.) 8–98) that binds BARD1,1 the product of another breast cancer susceptibility gene,39 which is required for the E3 ubiquitin ligase activity of the BRCA1-BARD1 heterodimer.40 More centrally, there is a coiled-coil motif from a.a. 1367–1437 that binds PALB2.41 42 PALB2 is also the product of a breast cancer susceptibility gene and is a key partner of BRCA2.43 44 Finally, there are two BRCT (BRCA1 C terminus) repeats, which mediate binding to Abraxas, BRIP1 and CtIP, from a.a. 1646–1859, very near the C-terminus of BRCA1.45 Interestingly, both BRIP1 and Abraxas are also breast cancer susceptibility proteins.39 46 47
BRCA2 has an N-terminal domain (a.a. 10–40) that binds to PALB2.44 More centrally, there are eight interspersed BRC motifs (a.a. 1008–2082) that bind the RAD51 recombinase.48 49 At the C-terminus, there is an additional RAD51 binding region (a.a. 3270–3305), and a helical domain (a.a. 2402–2668) and three OB folds that bind to single-strand DNA (a.a. 2670–3102), which are important for BRCA2 function.50–52 Recently, an additional DNA-binding domain (DBD) in BRCA2 has been identified between a.a. 250 and 500.53
As a basis for understanding functional assays for BRCA1 and BRCA2 variants, we will begin with a brief overview of the biological roles of these tumour suppressors in cells. BRCA1 has been reported to function in such diverse processes as transcriptional regulation and the control of centrosome number.42 54 55 It also has multiple roles in the cellular response to DNA damage, including regulation of the G2 DNA damage checkpoint,55 protection of stalled replication forks against nucleolytic degradation,56 mediating resistance to ionising radiation (IR) and chemotherapeutic agents,57 58 and mediating DNA repair by HR.59
Most of the properties of BRCA1 described above have been the basis for functional assays for BRCA1 variants at some point in time.22 These studies are catalogued on the BRCA1 Circos Web tool.60 Assays based on HR or cellular resistance to DNA damage have been used most recently and may be the most promising.34 35 38 61 One reason for this is that these assays have the potential to assess variants located throughout different regions of BRCA1, and therefore have greater general utility. In contrast, while transactivation by BRCA1 may also be used for functional assays,22 this is only relevant for VUS located in the extreme C-terminus of BRCA1 where this activity is located.54 62
Specifically, the function of BRCA1 in HR and cellular resistance to DNA damage requires the RING domain that has a role in the recruitment of BRCA1,34 36 61 a region from a.a. to 1–324 that interacts with the MRE11-RAD50-NBS1 complex to promote end resection,63 and the coiled-coil for recruitment of PALB2-BRCA2-RAD51 to initiate strand invasion.41 45 64 Additionally, the BRCT repeats are necessary for recruitment of BRCA1 and for binding to the CtIP nuclease as part of end resection.36 65 Thus, nearly any truncation or variant affecting BRCA1 protein stability inhibits HR. It should also be noted that HR assays and DNA damage sensitivity assays are readily quantifiable, which is highly desirable in functional assays.
BRCA2 also has a role in transcriptional activation, and functions in cytokinesis, the maintenance of G2 checkpoint arrest, the protection of stalled replication forks against degradation, and as a mediator of HR.45 56 66–68 Specifically, BRCA2 regulates the recruitment of RAD51 to sites of DNA damage and the oligomerisation of RAD51 with single-strand DNA that is necessary to form recombination intermediates.45 69 70 Functional assays based on direct measures of HR or on BRCA2-dependent assembly of RAD51 foci are particularly well suited to assessing BRCA2 VUS. BRCA2 also has a clear role in mediating cellular resistance to DNA damage.49 52 Correspondingly, BRCA2 functional assays have focused on HR using reporters and measurements of resistance to DNA damage.33 71–73 Consistent with this, the function of BRCA2 as a tumour suppressor is attributed to its central role in the DNA damage response.45
The use of HR or DNA damage sensitivity assays to assess BRCA2 VUS is also supported by the fact that pathogenic HBOC-associated missense variants of BRCA2 in the N-terminal PALB2-binding domain or the C-terminal DBD disrupt these processes. In comparison to transactivation by BRCA2, which involves only a small domain from a.a. 18–105,66 HR and resistance to DNA damage require domains throughout BRCA2. Thus, assays related to these functions of BRCA2 may hold particular promise as a means to assess variants distributed throughout the protein.33 45 70–73 It does not appear, however, that assays based on replication fork stability, which is related to DNA repair, accurately predict the cancer risk associated with BRCA2 VUS; the pathogenic Y3308X mutant promotes fork stability.72 74
Review of functional assays for BRCA1 VUS that use the full-length protein
As discussed in the previous section, assays based on HR and/or resistance to DNA damaging agents have emerged as the standard for the functional characterisation of BRCA1 VUS. Since multiple domains of BRCA1 are required for HR and resistance to DNA damage, assays with fragments of BRCA1, such as a recent assay based on the ubiquitin-ligase activity of the RING domain,37 may not be as accurate as assays that use the full-length protein. Thus, we will focus here on cell-based functional studies that have examined BRCA1 variants in the context of the full-length protein. Overall, the four studies of BRCA1 VUS discussed here have been validated as reliable for the RING and BRCT domains.34 35 38 61
Validation of functional assays utilising neutral and deleterious standards is necessary to establish the particular system that is employed.22 The studies we consider here have all been validated with such standards. The first such assay system established for BRCA1 was based on rescue of proliferative defects resulting from deficiency of mouse embryonic stem cells (ESCs) for endogenous Brca1 and employed heterologous expression of human BRCA1 in ESCs.35 Two missense variants of the BRCT repeats were examined, along with one neutral missense variant and three deleterious variants (two frameshift/one missense). Whether rescue of a proliferative defect, in the absence of exogenous DNA damage, is highly relevant to cancer susceptibility is uncertain.
Subsequently, another study in mouse ESCs characterised 74 missense BRCA1 VUS.61 To date, this is the largest individual functional study of BRCA1 VUS performed using the full-length protein. One reason for the relatively greater capacity of this assay was the use of a BRCA1 cDNA, encoding wild-type BRCA1 and variants, rather than a BAC to carry BRCA1. Still, this approach may not be rapid enough for a higher throughput analysis. Three neutral missense variants and five deleterious variants (two missense/three frameshift) were used as controls. All VUS found to be deleterious failed to confer resistance to cisplatin, and were restricted to the RING domain and the BRCT repeats. A subset of these missense VUS was also examined by measurements of HR using a reporter assay. Predictions based on HR and resistance to cisplatin agreed for all variants except R1699Q. R1699Q was defective for resistance to cisplatin but showed differences in HR that were not significantly different from the activity of wild-type BRCA1. Family-based studies have characterised R1699Q as an intermediate breast and ovarian cancer risk, which may explain the discordant functional results and underscores the need to include segregation analyses whenever possible.75
Systems to test BRCA1 and BRCA2 VUS in ESCs, including those described above, typically have constitutive knockout of one allele and conditional deletion of the other following introduction of human BRCA1 or BRCA2. Notably, due to multiple genetic manipulations and selection steps, these systems are labour-intensive and relatively slow, and therefore likely may not have the capacity to analyse the number of variants necessary to have an important impact.
BRCA1 has 58% conservation between humans and mice.76 As shown in figure 3, conservation is often high at the N-termini and C-termini of BRCA1, which contain the RING domain and BRCT repeats, respectively. It should be noted that pathogenic missense variants of BRCA1 in ClinVar reside predominantly in these RING and BRCT repeats. Importantly, systems based on heterologous expression of human BRCA1 in mice may not accurately assess variants located in domains with lower levels of conservation. It is also unclear whether mouse ESCs are a reliable model for preneoplastic events that increase the risk of developing cancer in human epithelial cells of the breast and ovaries.
Two additional studies characterising BRCA1 variants in HeLa cells, depleted of endogenous BRCA1 utilising an RNAi, have been conducted by the laboratory of Jeffery Parvin.34 38 In the first of these studies,34 five deleterious and one neutral variant, all missense, along with nine N-terminal VUS, all present in the first 71 amino acids of BRCA1, were examined based on measurements of HR. Interestingly, deletions of a.a. 1–302, 305–770, 775–1292 and 1527–1863 all compromised HR to some degree, further suggesting that different regions of BRCA1 are all required for HR or protein stability. The second study examined seven pathogenic and one non-pathogenic missense variants, and four missense VUS present from a.a. 90–170.38 Notably, a comparison of HR and another DNA repair process, single-strand annealing (SSA), showed that HR may more accurately distinguish previously classified benign and pathogenic variants. Thus, HR is a more reliable assay for the classification of BRCA1 VUS than SSA. While these assays were conducted in human cells, there are, nevertheless, certain concerns about them. Since HeLa cells are already transformed, they may not accurately model the role of compromised HR activity of BRCA1 in tumour initiation. Further, variable depletion of BRCA1 using RNAi could potentially make it difficult to compare the results of functional assays conducted in different laboratories. Certain VUS overlapped in the four studies described above, resulting in collective characterisation of 86 distinct missense variants.
Review of functional assays for BRCA2 VUS that use the full-length protein
Given how BRCA2 is believed to function as a tumour suppressor, assays related to DNA repair are directly relevant to predicting the impact of BRCA2 variants on cancer risk and therapeutic response. Additionally, such assays have demonstrated high sensitivity and specificity for predicting known benign and pathogenic variants. For these reasons, DNA repair-related assays are considered here.21
Since DNA repair-related domains are distributed throughout BRCA2, as discussed earlier, such assays should be based on expression of full-length BRCA2. BRCA2 is even larger than BRCA1 (the protein is ~390 kDa and the cDNA is 10 254 bp). Thus, it has been difficult to express full-length BRCA2 in human cells using a cDNA.69 73 As such, some functional studies of BRCA2 VUS have been based on heterologous expression of full-length BRCA2 variants in mouse ESCs using BACs.71 72 These studies, in the laboratory of Shyam Sharan, focused on VUS in the N-terminal PALB2-binding domain and the C-terminal DBD, where conservation between mouse and human BRCA2 is generally high (figure 4). Less than 10 distinct missense BRCA2 VUS were examined in each of these studies for the capacity to rescue proliferative defects resulting from conditional deletion of Brca2. 71 72 These studies employed one neutral missense and three deleterious variants (one missense/two frameshift),72 and two benign and one neutral missense variants, respectively.71 Importantly, results with benign and pathogenic standards corresponded precisely in the ability to rescue proliferative defects, to promote HR, measured utilising a reporter assay or based on the assembly of RAD51 foci, and to promote resistance/sensitivity to DNA damage. Although studies with larger numbers of variants are needed as a basis for more definitive conclusions, this suggests the possibility that these assays may be used interchangeably to classify BRCA2 VUS.
Two studies led by the laboratory of Fergus Couch heterologously expressed full-length human BRCA2 in BRCA2-deficient VC-8 hamster lung fibroblast cells.33 73 They began their studies by transiently expressing BRCA2 in human cells, but had difficulty achieving efficient expression.73 Since the reliability of assays and the ability to compare results from independent experiments are greatly facilitated by efficient and stable expression, the investigators ultimately turned to cDNA-based expression in VC-8 hamster cells. In one of these studies, they tested one nonsense variant which they defined as neutral and one frameshift mutation they defined as deleterious for resistance to mitomycin C (MMC) or IR.73 They also tested seven BRCA2 VUS for resistance to DNA damage, most of which were additionally assayed for activities in HR and suppression of centrosome amplification. All but one variant, T2515I, gave concordant results between these different assays. T2515I was deficient for resistance to DNA damage but was intermediate for HR and centrosome amplification. This again emphasises the importance of assay selection and underscores the need for setting cut-offs to distinguish variants associated with no, partial or full loss of function. In a subsequent study by this same group,33 13 pathogenic/likely pathogenic and 17 non-pathogenic/likely non-pathogenic missense standards were used to set cut-offs for interpreting the results for 33 missense VUS of the C-terminal DBD of BRCA2.
When overlap is accounted for, the above studies conducted using full-length BRCA2 together considered 54 missense VUS. Therefore, only 86 and 54 missense VUS of BRCA1 and BRCA2, respectively, have been functionally characterised. Assays with a greater capacity must therefore be developed to keep pace with demand, since ~1300 and ~2300 distinct missense VUS of BRCA1 and BRCA2, respectively, are currently listed in ClinVar.
Another issue with the studies of BRCA2 VUS described above is that they were all based on heterologous expression in rodent cells. Mouse and human BRCA2 have 59% conservation.77 As examples of issues raised by inexact conservation, non-conservative changes two and one a.a., respectively, C-terminal to the IARC class 5 (pathogenic) R3052W variant and the class 4 (likely pathogenic) L2647P variant, both of which are present in the relatively highly conserved C-terminal DBD of BRCA2, are shown in figure 4B. Two other class 5 pathogenic variants present in this domain, W2626C and I2627P, of eight proposed as part of a validation panel for functional studies of BRCA2 VUS,21 have non-identical amino acid residues within two positions of the variant residue (not shown). The fact that inexact conservation can potentially affect pathogenic variants is important because this could lead to inappropriate classification.
Heterologous assays of BRCA2, as well as BRCA1, may have particular difficulty accurately assessing variants in areas of lower level conservation. As an example, here we consider the case of BRCA2. Of the 72 IACR class 1 neutral variants proposed for the validation of functional studies,21 60 are outside the highly conserved PALB2-binding and DBD domains. For 34 of these 60 neutral variants (57%), the variant residue does not align with the mouse sequence (including H1918Y, L2396F, N2436I, K2472T) or the corresponding residue is not conserved in mice (including N56T, K513R, R1190W, C1265S and G1771D). An example is shown for K2472T (figure 4C), which lacks alignment with corresponding residues in mouse BRCA2 for a.a. 2460–2478, suggesting the possibility that a subdomain in which this variant resides may not be present in mouse BRCA2. Additionally, K513R provides an example where there is a non-conservative change in the mouse sequence corresponding to the position of the variant, flanked by non-conserved residues. Such lack of conservation is a potential issue since many of the functions of BRCA2, as well as BRCA1, are mediated by protein interactions. Interacting proteins in rodent cells may not interact as efficiently with human BRCA1 or BRCA2 in domains with low conservation. The consequence of this is that certain domains of human BRCA1 or BRCA2 may not have their normal activities in rodent cells due to low conservation. Thus, the effect of variants in these domains may be underestimated or not measured at all. Therefore, assays in human cells are instead necessary to assess cancer risk, as well as increased sensitivity to chemotherapy for the many BRCA1 or BRCA2 variants (shown in figure 3,4) that are present outside of highly conserved regions. Again, while most pathogenic variants identified so far reside in highly conserved domains, the ability to functionally characterise variants in other areas is necessary to fully empower genetic screens of BRCA1/2.
Assessment of what is needed to drive more efficient and reliable classification of BRCA1/2 VUS
Essential controls for functional assays
Functional assays to characterise BRCA1 and BRCA2 missense VUS need positive and negative controls to validate the sensitivity and specificity of the assay. Missense variants that have been clinically classified as benign should behave as wild-type and those classified as pathogenic should behave similar to knockout cells or truncating variants. Controls should include those that have been classified using robust multifactorial models and/or for which the significance has been evaluated with genetic evidence such as segregation.
What functional assays are most relevant for predicting pathogenicity?
Among the many functions of BRCA1 and BRCA2, HR may be especially important for preventing cancer susceptibility. In most studies of both BRCA1 and BRCA2 VUS, HR is a highly sensitive and specific assay for missense variants, as discussed in the two previous sections. In comparison, while sensitivity to PARP inhibitors may have a reasonably high-throughput capacity and is highly relevant for guiding treatment, it may not be as relevant or accurate as HR for predicting cancer risk. For example, in a mouse model, the pathogenic BRCA1 C61G variant, which is well-associated with HBOC and causes tumours in mice, does not appear to render the tumours sensitive to platinum drugs or PARP inhibition.78
Tissue and model specificity
A relatively unaddressed issue in the development of optimal functional assays for BRCA1/2 is whether tissue specificity matters. BRCA1 and BRCA2 are important for DNA repair across tissue types. However, there is tissue specificity for which organs and cells of origin are at highest risk for developing tumours in carriers of these mutations. Thus, there are likely tissue-specific factors that may modulate the context in which these mutations influence cancer development. There has been no direct side-by-side comparison of variant assays from breast/ovarian cells to cell lines derived from other tissues. Similarly, functional assays for BRCA1/2 have been done in cells from organisms such as mouse ESCs, yeast and hamster.33 35 61 71–73 79 Aside from issues of inexact conservation, there may be subtle differences between human and other organisms that could impact results.
Improved systems for cDNA-based expression of BRCA1 or BRCA2 in human cells with a corresponding deficiency
Expression of full-length BRCA1 or BRCA2 is difficult, especially for BRCA2, and this has led to the use of heterologous expression systems. Additionally, there are few BRCA1-deficient and BRCA2-deficient human cell lines available for assays,80–83 and not necessarily in the most relevant tissue type or with the appropriate transformation/immortalisation status. Overcoming these challenges may require innovative approaches to gene editing and gene expression. Such innovation, however, should lead to the capacity to conduct higher throughput assays using cDNA-based approaches in human tissue-specific models that better predict cancer risk or, separately, therapeutic response.
Incorporation of data from functional assays into multifactorial analyses
A final important consideration is how best to incorporate data from functional assays for BRCA1 and BRCA2 into multifactorial predictive models. Statistical algorithms calculating the probability of pathogenicity can be developed using the variances of distribution of a quantitative functional assay along with the means and confidence intervals (CIs) of each variant being studied, including variants of known classification. A pathogenicity probability based on these can be used to calculate likelihood ratios. Likelihood ratios can be integrated with other available factors in the model to provide an overall likelihood probability of pathogenicity for a variant (figure 5). One example of this is VarCall, a computational tool that incorporated functional data from the C-terminal domain of BRCA1 to determine the likelihood of pathogenicity for over 200 missense variants.84 The power of predicting pathogenicity of missense variants in this domain increased dramatically, suggesting that this type of computational algorithm may be quite useful once large amounts of robust functional data are available.
Conclusion
Collectively, these studies show the power of functional assays for predicting pathogenicity of BRCA1/2 VUS. Moving forward, it will be important to define the optimal assays to use for BRCA1/2 VUS classification, which can be incorporated into multifactorial statistical analysis models that also include any available data on cosegregation and tumour pathology.
References
Footnotes
Contributors AET and PRA both contributed to conceptualisation and writing of the manuscript.
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.