Article Text
Abstract
Background Mitochondrial disorders are associated with abnormalities of the oxidative phosphorylation (OXPHOS) system and cause significant morbidity and mortality in the population. The extensive clinical and genetic heterogeneity of these disorders due to a broad variety of mutations in several hundreds of candidate genes, encoded by either the mitochondrial DNA (mtDNA) or nuclear DNA (nDNA), impedes a straightforward genetic diagnosis. A new disease gene is presented here, identified in a single Kurdish patient born from consanguineous parents with neonatally fatal Leigh syndrome and complex I deficiency.
Methods and results Using homozygosity mapping and subsequent positional candidate gene analysis, a total region of 255.8 Mb containing 136 possible mitochondrial genes was identified. A pathogenic mutation was found in the complex I subunit encoding the NDUFA9 gene, changing a highly conserved arginine at position 321 to proline. This is the first disease-causing mutation ever reported for NDUFA9. Complex I activity was restored in fibroblasts of the patient by lentiviral transduction with wild type but not mutant NDUFA9, confirming that the mutation causes the complex I deficiency and related disease.
Conclusions The data show that homozygosity mapping and candidate gene analysis remain an efficient way to detect mutations even in small consanguineous pedigrees with OXPHOS deficiency, especially when the enzyme deficiency in fibroblasts allows appropriate candidate gene selection and functional complementation.
- NDUFA9
- complex I
- Leigh syndrome
- homozygosity mapping
- candidate gene identification
- genetics
- metabolic disorders
- molecular genetics
- muscle disease
- neuromuscular disease
- cell biology
- academic medicine
- neuroophthalmology
- cardiomyopathy
- diagnostics tests
- arrhythmias
- congenital heart disease
Statistics from Altmetric.com
- NDUFA9
- complex I
- Leigh syndrome
- homozygosity mapping
- candidate gene identification
- genetics
- metabolic disorders
- molecular genetics
- muscle disease
- neuromuscular disease
- cell biology
- academic medicine
- neuroophthalmology
- cardiomyopathy
- diagnostics tests
- arrhythmias
- congenital heart disease
Introduction
Mitochondrial disorders are clinical phenotypes associated with abnormalities of the oxidative phosphorylation (OXPHOS) system. OXPHOS disorders are among the most common inherited metabolic disorders, with a prevalence of 1 in 5000–8000, and cause significant morbidity and mortality in the population.1 They can present at any age and in any tissue. A broad range of clinical features has been associated with defects in the mitochondrial DNA (mtDNA) and in nuclear genes encoding essential proteins involved in the OXPHOS system.2 3 Because of this dual genetic control, segregation of OXPHOS disorders can be complex with maternal, autosomal recessive, autosomal dominant or X linked inheritance patterns. Mutations in nuclear mitochondrial genes include genes encoding assembly factors, complex subunits of the OXPHOS system, and genes involved in mitochondrial maintenance or metabolism in general.4 Mutations in different genes can lead to similar phenotypes, while mutations in the same gene can give rise to different phenotypes, illustrating the clinical and genetic heterogeneity of these diseases. Nuclear genes are likely the major cause of mitochondrial disease, especially in affected children, and, based on the mitochondrial proteome, potentially more than 1500 genes could be involved.5 Pathogenic mutations have been identified in ∼100 genes during the past years. However, mutations are often limited to only one or a few patients and families. The genetic cause still remains unknown in a large proportion of patients with mitochondrial disease.6
Several approaches exist to identify gene defects in patients with mitochondrial disease, such as conventional sequencing of candidate genes,5–7 family based approaches,6 8–10 and next generation sequencing.11–13 Consanguinity or suspected relatedness of the parents of OXPHOS patients makes homozygosity mapping followed by candidate gene analysis a rapid and effective approach. Here we report the first pathogenic mutation in NDUFA9 in a patient from consanguineous parents with fatal Leigh syndrome and complex I deficiency using this approach. Subsequently, the pathogenicity of the mutation was confirmed by complementation studies in complex I deficient fibroblasts of the patient. The procedure from the initial single nucleotide polymorphism (SNP) array analysis until the identification of the mutation took about 3 weeks, indicating that this type of approach is efficient in terms of success rate, time, and costs.
Subjects and methods
Clinical findings
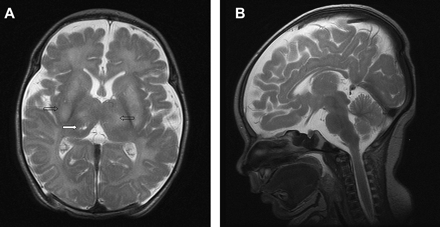
The patient was a boy born as the first and only child from consanguineous Kurdish parents (second cousins, region Kurdistan in Iraq) at 41 weeks of gestation with a birth weight of 3.040 kg (−1 SD) and an occipitofrontal circumference (OFC) of 33.8 cm (−1 SD). No dysmorphic features were noted. A mildly depressed child was seen at birth with intact functioning of the cranial nerves and a normal somatic motor and sensory system with symmetric reflexes. After birth, the child deteriorated and suffered from a combined respiratory and metabolic acidosis with increased lactate values up to 10 mM 12 h postpartum (hyperlactataemia >2.5 mM). A metabolic investigation in blood and urine revealed normal amino acid values, a slightly decreased total carnitine value with a slightly elevated alanine value and an increased lactate/pyruvate ratio (>20). In urine, lactic aciduria was present. Morphological investigations in muscle, including electron microscopy, did not reveal any abnormalities. Mitochondrial oxygen consumption was decreased and an isolated complex I deficiency was present in muscle (biopsy taken from right quadriceps muscle) and fibroblasts (respectively, 29% and 11% activity compared to control values). The boy developed profound hearing loss, apnoeas associated with brainstem abnormalities, and retinitis pigmentosa. A brain MRI at day 6 showed diffuse loss of supratentorial white matter and brain stem volume with T2 hyperintensities of the basal nuclei (thalamus and putamen) and a right-sided focal thalamic lesion compatible with striatal ischaemia and focal necrosis, leading to a diagnosis of Leigh syndrome (figure 1). Electroencephalograms (EEGs) taken at 1 day, 1 week, 2 and 3 weeks of age showed increased aberrations of the normal pattern with multifocal sharp waves and a discontinuous pattern. No overt epileptic seizures were seen. His legs became increasingly hypertonic with choreadystonic movements from all limbs. At 3 weeks he needed ventilatory support. He died 1 month after birth due to respiratory insufficiency. Deletions and mutations in the mtDNA were excluded as a cause of disease by, respectively, long range PCR and the Affymetrix Resequencing Mitochip 2.0 in DNA isolated from muscle as described previously.14
Brain MRI leading to the diagnosis of Leigh syndrome. (A) Axial T2 weighted image shows high signal intensity in the basal ganglia and thalami bilaterally (open arrows). In the right thalamus, signs of striatal necrosis can be seen (closed arrow). The cerebral white matter is hyperintense. (B) Sagittal T2 weighted image, showing diffuse cerebral and cerebellar atrophy.
Homozygosity mapping
Homozygosity mapping was performed with the Affymetrix GeneChip Human Mapping 250 K NspI SNP array using peripheral blood DNA of the patient. The DNA was processed and labelled according to the instructions of the manufacturer. Genotypes were generated by the Affymetrix GeneChip Genotyping Analysis Software (GTYPE). For the identification of homozygous regions the online tool ‘HomozygosityMapper’ was used.15 Candidate regions were selected in which at least 400 consecutive SNPs were homozygous in the patient.
Sequence analysis
Exons and flanking intronic regions of 40nt from the NDUFA9 gene were PCR amplified using intronic primers (Biolegio, online supplementary table 1). The other candidate gene associated with complex I, NDUFB2, was not sequenced after the pathogenic NDUAF9 mutation was identified. PCR products were directly sequenced with the PRISM Ready Reaction Sequencing Kit (Perkin-Elmer Life Sciences, Boston, MA, USA) on an ABI3730 automatic sequencer (Applied Biosystems, Foster City, CA, USA).
Blue native (BN)-polyacrylamide gel electrophoresis and western blotting
Mitoplasts from patient and control fibroblasts were isolated as previously described.16 Mitochondrial protein (15 μg) were analysed on polyacrylamide gradient gels (4–16%; Invitrogen, Carlsbad, CA, USA) for the Blue native. For western blot analysis 20 μg of total protein was used, and to visualise the specific proteins, monoclonal antibodies were used raised against complex I subunits NDUFA9, GRIM19, NDUFB8 and 70 kDa complex II subunit (Mitosciences, Eugene, OR, USA).
Immunofluorescence staining
Fibroblasts were fixated with 4% paraformaldehyde followed by permeabilisation with 0.25% Triton X100. A NDUFA9 antibody (Mitosciences) was used to stain for complex I and visualised using an Alexa Fluor® 488 secondary antibody (Invitrogen).
Complementation assay
An FIV (feline immunodeficiency virus) based lentiviral system was used to transduce patients and control cell lines with wild type and mutant NDUFA9, as described previously.7 10 The activities of complex I and citrate synthase were measured spectrophotometrically as described by Sgobbo et al.17
Results
Homozygosity mapping
In the patient, 32 regions with at least 400 consecutive, homozygous SNPs were present on 14 different chromosomes ranging from 3.8 Mb to 21.1 Mb with a total of 255.8 Mb. Genes with a possible mitochondrial function were identified using continuously updated information from the MitoP2 database,18 19 the MitoCarta compendium20 21 and our own literature based information. Of the 136 candidate genes identified in the homozygosity regions, two genes were known to be related to complex I: NDUFB2 (NADH dehydrogenase (ubiquinone) 1 β subcomplex, 2, 8 kDa), and NDUFA9 (NADH dehydrogenase (ubiquinone) 1 α subcomplex, 9, 39 kDa).
Mutation analysis revealed a p.R321P mutation in NDUFA9
In the patient a homozygous c.962G>C substitution was identified in NDUFA9 leading to an amino acid change from arginine to proline (p.R321P). Both parents were heterozygous for the substitution. The amino acid at position 321 is highly conserved from human to Drosophila melanogaster (figure 2). The substitution was predicted to be damaging to protein function by the programs SIFT22 and PolyPhen,23 based on evolutionary conservation. Analysis with SOPM,24 PREDATOR25 and PSIPRED,26 which use sequence information and similarities to predict secondary protein structures, showed that the secondary structure changed due to loss of an α helix in the mutant protein. The NDUFB2 gene was not sequenced anymore after the pathogenic NDUAF9 mutation was identified.
Amino acid alignment of the human NDUFA9 reference sequence versus various species for position 301 to 346. Indicated is the position of the novel p.R321P mutation. The arginine at position 321 is conserved in all species shown, except for Caenorhabditis elegans.
Wild type NDUFA9 restores complex I deficiency in patient fibroblasts
Complex I measurements in fibroblasts of patients, transduced with lentiviral clones containing wild-type or mutant NDUFA9 showed that only wild type NDUFA9 was able to restore the complex I activity in the patient fibroblasts, confirming that the p.R321P mutation causes the complex I deficiency and related disease (table 1).
Restoration of complex I activity after complementation in fibroblasts with the p.R321P mutation in NDUFA9
To investigate if the reduced complex I activity in patient fibroblasts was due to a decrease in the total amount of complex I, Blue Native PAGE was performed on mitoplasts. Results using antibodies against GRIM19 and NDUFB8 demonstrated a strong decrease in the amount of mature complex I, which was only barely visible (figure 3). Transduction with wild type NDUFA9 resulted in an increased amount of complex I, which is compatible with the restored complex I activity (table 1). A western blot was performed on total protein extract from fibroblasts to check whether the mutation affected protein stability. A decrease in total amount of NDUFA9 protein was observed in the patient (figure 4), which could be restored by transduction with wild type NDUFA9 (data not shown). Also, a decreased amount of GRIM19 and NDUFB8 protein was observed (figure 4). Immunofluorescence staining using anti-NDUFA9 only showed a signal in wild-type cells and in the patient cell line complemented with wild-type NDUFA9 (figure 5).
Total amount of mature complex I is restored in fibroblasts of the patient transduced with WT NDUFA9. BN-PAGE followed by western blotting using the anti-GRIM19 and the anti-NDUFB8 antibody shows a decrease in the amount of complex I compared to complex II in patient versus control fibroblasts. Complementation of patient fibroblasts with wild-type NDUFA9 (Pwt), but not with mutant NDUFA9 (Pmut) restores the amount of complex I. Cwt: complementation of control with wild-type NDUFA9; Cmut: complementation of control with mutant NDUFA9.
Western blot analysis on total protein extract from fibroblasts shows instability of the NDUFA9 protein. Results show a decreased amount of NDUFA9, GRIM19, and NDUFB8 protein in the patient using anti-70 KDa complex II subunit as a control.
Immunofluorescence staining shows absence of complex I (NDUFA9) in fibroblasts of the patient. Red fluorescence represents mitochondria, green fluorescence complex I, and blue fluorescence the nucleus. In the patient, complex I is absent as shown in the middle row and in the merged pictures. Complementation with wild-type NDUFA9 in patient fibroblasts clearly restores the amount of complex I protein.
Discussion
Our data show that NFUFA9 defects are a new cause of Leigh syndrome. We report the first mutation in NDUFA9 identified in a child with Leigh syndrome and complex I deficiency from consanguineous parents, changing a highly conserved arginine at position 321 by proline. Complex I activity was restored in fibroblasts of the patient by lentiviral transduction with wild-type but not with mutant NDUFA9. These data confirm that the NDUFA9 mutation causes the complex I deficiency and related disease. Defects in the mtDNA as well as in nuclear genes have been described in patients with complex I deficiency and Leigh syndrome.8 Mutations have been identified in several essential complex I genes in Leigh syndrome, like NDUFV1,27 NDUFS1,28 NDUFS3,29 NDUFS4,30 NDUFS7,31 NDUFS8,32 NDUFA1,33 NDUFA2,34 NDUFA10,35 NDUFAF2,36 c8orf38,21 c20orf7,8 and FOXRED1.11 Most of these genes encode for subunits belonging to the core subunits of complex I, while others are accessory subunits or assembly factors. The exact role of FOXRED1 in complex I is still unclear.
NDUFA9 is one of the nuclear encoded structural accessory subunits located in the peripheral part of complex I. Previous findings have shown that absence of the NDUFS8 subunit, which is thought to connect the membrane and peripheral domains of complex I,37 led to dramatically reduced levels of NDUFA9.38 This suggested that NDUFA9 serves as an anchor connecting the peripheral arm with the membrane arm.37–39 We hypothesise that the structurally altered NDUFA9 in the patient is not properly incorporated, resulting in loss of this connection. This may lead to the instability and fast degradation of the complex I intermediates and almost complete absence of the mature complex I. Seven of the 45 complex I subunits are mtDNA encoded while the remaining 38 are nDNA encoded.40 The latter group consists of seven core subunits and 31 accessory subunits. For the core subunits, primary roles in electron transfer and proton translocation have been demonstrated, while the exact role of the accessory subunits is less well understood. A function in the stabilisation or biogenesis of the enzyme complex has been proposed, but also regulation of activity or the assembly of other subunits into the holocomplex as well as a role in preventing ROS generation and protection against oxidative damage.35 41 42 Our data indicate an important role for NDUFA9 in proper complex I function, probably due to its role in maintaining stability.
In our cohort, no patients with a similar phenotype with complex I deficiency and Leigh syndrome were present. However, we have additionally screened the NDUFA9 gene in 34 patients with complex I deficiency (ranging from 13–70% complex I activity of the controls), in whom no mutations were present in the mtDNA. None of these patients was diagnosed with Leigh syndrome. In other patients with Leigh syndrome available for screening, Leigh syndrome was associated with a deficiency of other mitochondrial respiratory chain complexes. No additional pathogenic NDUFA9 mutations were identified, confirming the emerging picture that nuclear gene defects in mitochondrial diseases are limited to only a few patients and families and demonstrating that Leigh syndrome is a genetically heterogeneous neurodegenerative disorder. This NDUFA9 defect causes a severe Leigh phenotype expressed by the early death of the child, and the combination of diffuse affection of all brain structures as seen with cerebral MRI and the striatal necrosis.
Massive parallel sequencing of candidate genes or all genes (exome) is increasingly and successfully applied to identify the underlying gene defect in patients with mitochondrial disease.12 13 Recent advances in sequencing technologies show that combining whole exome sequencing with homozygosity mapping is a successful approach to identify disease loci in patients.43 44 It can be expected that with the increase in speed and drop in costs of these approaches this method will become the method of choice, especially when the number of candidate genes is large and the selection criteria limited. As these new approaches will yield increasing numbers of potentially damaging variants, it will be essential to incorporate functional tests to distinguish the disease-causing mutations from other variants. Our study shows that homozygosity mapping and candidate gene analysis can still be a successful alternative in selected patients and that in our hands the lentiviral transduction system is a rapid and effective way for performing complementation assays in OXPHOS disease.
Acknowledgments
We are grateful to Dr M Vermeulen (paediatrician at the Department of Pediatrics, Erasmus MC, Rotterdam) for referring the patient to us, and Dr G Mancini for genetic counselling of the family. SNP-array analysis was performed by the section Cytogenetics of the Department of Human Genetics (Nijmegen, The Netherlands). We would like to thank Dr G Nicolaes for performing the prediction analysis using PREDATOR and PSIPRED.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Files in this Data Supplement:
- Download Supplementary Data (PDF) - Manuscript file of format pdf
Footnotes
BJCB and MG contributed equally to this work.
Funding This work was supported by an EU grant to the MitoCircle project (Sixth Framework Program, contr. no. 005260) and by the Alma in Silico project (EMR. INT4.-1.3.-2008-03/003). HP was supported by the Impulse & Networking Fund of the Helmholtz Association in the framework of the Helmholtz Alliance for Mental Health in an Ageing Society (HA-215), the German Network for Mitochondrial Disorders (mitoNET 01GM0862 and 01GM0867), and Systems Biology of Metabotypes (SysMBo 0315494A).
Competing interests None.
Patient consent Obtained.
Ethics approval This study was performed within the diagnostic patient care at the Department of Clinical Genetics.
Provenance and peer review Not commissioned; externally peer reviewed.