Article Text
Abstract
We describe a family with non-syndromic sensorineural hearing impairment inherited in a manner consistent with maternal transmission. Affected members were found to have a novel heteroplasmic mtDNA mutation, T7510C, in the tRNASer(UCN) gene. This mutation was not found in 661 controls, is well conserved between species, and disrupts base pairing in the acceptor stem of the tRNA, making it the probable cause of hearing impairment in this family. Sequencing of the other mitochondrial tRNA genes did not show any other pathogenic mutations. Four other mutations causing hearing impairment have been reported in the tRNASer(UCN) gene, two having been shown to affect tRNASer(UCN) levels. With increasing numbers of reports of mtDNA mutations causing hearing impairment, screening for such mutations should be considered in all cases unless mitochondrial inheritance can be excluded for certain.
- hearing impairment
- mtDNA mutation
- tRNASer(UCN)
Statistics from Altmetric.com
Sensorineural hearing loss is the most common phenotype of mitochondrial diseases and to date four point mutations of the mitochondrial DNA (mtDNA) have been reported in families with non-syndromic, maternally inherited hearing impairment (MIHI). The most common, the A1555G mutation in the 12S rRNA gene (MIM 561000), also causes increased sensitivity to the ototoxic side effects of aminoglycosides.1 2 The other three, A7445G, 7472insC, and T7511C, all lie in the tRNASer(UCN)gene (MIM 590080) and in some cases hearing impairment can also be associated with other features, such as palmoplantar keratoderma or ataxia.3-6
We describe a white family with non-syndromic sensorineural hearing impairment transmitted in a manner consistent with maternal inheritance (fig 1A), that is, there was no paternal transmission and II.2 had hearing impaired children by two different fathers. The proband (IV.6) was formally diagnosed as having sensorineural hearing impairment aged 15 months. His hearing loss is profound but asymmetrical, with an average loss in the better ear of 103 dB HL (associated with a gently sloping audiogram profile) and 115 dB HL in the other ear. His younger sister (IV.7) was not formally diagnosed as having a hearing impairment until 5 years of age. Her sensorineural hearing loss is severe but asymmetrical with an average loss in the better ear of 66 dB HL (with a gently sloping audiogram profile) and 75 dB HL in the other ear.
(A) Pedigree of the family. Solid symbols denote those with hearing impairment. (B) Gain of HinfI restriction enzyme site in affected subjects. mtDNA was amplified with primers corresponding to bp 7392-7410 (forward) and 7608-7588 (reverse) and the PCR fragment digested to completion with 10 units of HinfI and fragments separated on a 2% agarose gel. The normal product gives fragments of 150 bp and 66 bp, the HinfI gain cuts the 150 bp fragment into 101 bp and 49 bp. Patient ID numbers correspond to those in (A), lanes 5 and 6 are hearing controls, and M is pBR322 HaeIII marker. (C) DNA sequence of the mtDNA heavy strand showing the T to C base change at bp 7510 in the proband compared to a hearing control. The proportion of mutant was estimated by densitometry of both the agarose and sequencing gels.
Their mother (III.9) had not previously been diagnosed as having a hearing loss but she was aware that her hearing was “dull at times”. Portable audiometry showed her to have a moderate sensorineural hearing loss, with an average loss in the better ear of 41 dB HL and 48 dB HL in the other ear.
The mothers' four sibs all have hearing impairment to varying degrees. The only other clinical feature of note in the family is mental handicap in a maternal aunt (III.7), which has previously been attributed to birth asphyxia. The maternal grandmother (II.2) died at the age of 80 and had suffered a loss of hearing for her last 10 years. With the exception of an uncle (III.5), who was only related by marriage, there was no history of exposure to ototoxic drugs or other illness in the family.
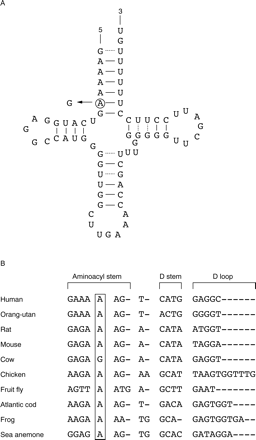
Following informed consent, DNA was extracted from blood taken from the proband, his sister, and both parents. No other family members were available for study nor were we able to carry out any further analysis of those available to us. Analysis by PCR and restriction enzyme digestion showed the absence of the A1555G, A7445G, T7511C, and 7472insC mutations. Further analysis, however, showed the gain of aHinfI site around base pair (bp) 7510 in the tRNASer(UCN) gene of the proband, his sister, and mother (fig 1B). Sequencing showed this to be a T to C transition at bp 7510 (fig 1C). The T7510C mutation was heteroplasmic in all three affected family members tested, that is, >95% mutant in the two sibs and 90% in the mother. The following points provide further support for the T7510C mutation as the most probable cause of the hearing impairment in this family. (1) The HinfI site gain at bp 7510 is extremely rare. We did not find it in 141 white controls here, nor was it reported in 520 other controls in published reports.7 8 (2) The base change disrupts a hydrogen bond in the acceptor stem of the tRNASer(UCN) (fig 2A), which may affect tRNA levels or function. Similar mutations in this acceptor stem at bp 7511 and 7512 have previously been shown to cause hearing loss.4 9 10 (3) The 7510 residue is highly conserved in a wide range of species (fig 2B) forming an A-U base pair in all except bovine.
(A) Proposed secondary structure of tRNASer(UCN). The gene is encoded on the light strand and thus the base change is shown as A to G in the tRNA. (B) Interspecies homology of mitochondrial tRNASer(UCN) genes. Aligned sequences were obtained from the tRNA compilation manual.17 Position 7510 is indicated by the boxed area.
Sequencing of the other mitochondrial tRNA genes from this family did not show any other changes except for a homoplasmic A to G base change at bp 4336 in the tRNAGln gene. This base change is present in about 1% of the white population and although it has been reported in patients with Alzheimer's or Parkinson's disease in some studies,7 any specific disease association remains uncertain.11 Hearing loss has not been reported in any of these persons, although such information was not actively sought. The A4336G mutation is therefore unlikely to be the primary cause of hearing loss in this family although it may contribute to the high penetrance of the T7510C mutation in this family.
Several mtDNA point mutations have been associated with non-syndromic sensorineural hearing loss. Generally these mutations are homoplasmic or present at very high levels (>95%) and presumably exert only very mildly deleterious effects which are sufficient to affect the inner ear but with no noticeable effect on other tissues. The reason for this tissue specificity is not known. Fischel-Ghodsian12 has suggested it could be because of different processing of the mitochondrial genes in the cochlea. Such a difference between tissues has been shown to exist, for example the A3302G mutation in the tRNALeu(UUR) gene13 causes a defect in RNA processing in skeletal muscle but not fibroblasts where there is no phenotypic defect.
It is interesting that five mutations in or affecting the tRNASer(UCN) gene (T7510C, T7511C, T7512C, A7445G, and 7472insC) have all been associated with hearing loss. Two of these, A7445G and 7472insC, have been shown to cause a significant reduction in tRNASer(UCN) levels and a mild mitochondrial defect in lymphoblasts or osteosarcoma cells.14 15 Whether or not the T7510C mutation has any affect on the processing of mitochondrial genes in the cochlea can only be answered by studying cells from the inner ear.
The increasing number of reports of families with hearing impairment resulting from mtDNA mutations, and the relatively high frequency of the A1555G mutation in Spain found by Estivill et al,16 suggest that mtDNA mutations may be a more common cause of familial non-syndromic hearing impairment than previously estimated. Sensorineural hearing impairment caused by mtDNA mutations is usually of childhood onset and progressive, although some persons may not be affected until much later in life. Given that some families with mitochondrial inherited hearing impairment may not be large enough to establish a pattern of maternal inheritance with certainty, screening for mtDNA mutations should always be considered in any family which does not exhibit paternal transmission of the hearing impairment.
Acknowledgments
This work was funded in part by a grant from Defeating Deafness. Work in the authors' laboratories is also supported by the Medical Research Council, European Community, Wellcome Trust, and Yorkshire Cancer Research. We are grateful to the family for their cooperation.