Abstract
The newly identified gene, ARX, when mutated has been shown to cause both syndromic and nonsyndromic forms of mental retardation. It seems that the less severe forms are due to polyalanine expansions and missense mutations in the gene. We screened 682 developmentally retarded males for polyalanine expansions in ARX in order to examine the contribution of ARX mutations to the causes of developmental retardation. We also reinvestigated 11 putative MRX and three MR families where no cause of mental retardation had been found, by mutational analysis of ARX. Mutational analysis was also performed in 11 probands with autism from families with two or more affected males. We find that previously described polyalanine expansions of ARX are not a common cause of mental retardation.
Similar content being viewed by others
Introduction
Mutations of the ARX gene have recently been identified as a major contributor to X-linked mental retardation (XLMR). The phenotypic expression varies, and mutations in ARX have been seen to cause nonsyndromic mental retardation1 as well as syndromic forms such as West syndrome (ISSX), Partington syndrome, infantile myoclonic epilepsy and spasticity2 and X-linked lissencephaly with abnormal genitalia (XLAG).3 Variation in phenotypic expression has also been described within a family.4 In XLMR families, ARX mutations have been suggested to be a more frequent cause of mental retardation than other XLMR genes.5
ARX maps to Xp22 and spans 12.5 kb of genomic DNA. It is composed of five exons encoding a protein of 562 amino acids. ARX belongs to the family of Aristaless-related paired-class homeobox genes. These genes are transcription factors and function as key players in vertebrate embryology. The Aristaless-related class of homeodomain proteins are characterized by a paired homeodomain (DNA binding) and an Aristaless domain (also called C-peptide, paired-tail or OAR domain). Besides these domains, a conserved octapeptide motif and four polyalanine tracts are present. The polyalanine tracts are not conserved throughout evolution and are not seen in zebrafish or Drosophila melanogaster. The ARX gene is otherwise conserved through evolution. ARX is expressed in the forebrain and in the floor plate of the developing CNS, the expression pattern depending on gestational age.1, 6
Two recurrent mutations, both leading to expansions of the polyalanine tracts, have been reported (c.431_454dup(24 bp) and c.333_334ins(GCG)7).1, 2 In addition, missense mutations, nonsense mutations and deletions/insertions leading to frameshifts and premature truncation have been reported.1, 2, 3 It seems that mutations leading to premature truncation of ARX generally cause the more severe XLAG phenotype.3
We have screened 682 males with nonspecific developmental delay/mental retardation, referred for fragile X DNA testing, by fragment size analysis of nucleotides 247–497 in order to detect the two recurrent mutations in ARX as well as other insertions/deletions in this area. Furthermore, we have sequenced the coding region plus flanking sequences of ARX in 25 probands from putative MRX, MR or autism families. We show that the previously reported polyalanine expansions in ARX are not a common cause of nonspecific mental retardation in Denmark.
Materials and methods
Patients
We selected 682 samples from males aged 2–75 years who were referred for fragile X testing from 1999 to 2002 and tested negative for a full mutation of FMR1. The males also tested negative for other chromosomal abnormalities as evidenced by standard chromosome analysis. These males were investigated for deletions and insertions in ARX from nucleotide 247 to 497 (numbering according to NM_139058).
In addition, the coding region and flanking sequences of ARX were sequenced in 11 putative MRX families, three MR families and 11 families with two or more affected male siblings with autism. The autistic children were diagnosed with infantile autism in child psychiatric departments according to ICD-10 criteria (WHO, 1992). Putative MRX families had three or more affected males plus an X-linked pattern of inheritance, while MR families had two affected males. Linkage analysis had been attempted in some of the families, but families were generally too small. In the family of case 29 442 linkage analysis was inconclusive.
Phenotypic description of patients with expansions and a deletion in ARX 29442
(c.431_454dup(24 bp))
This boy was born as number 2 to healthy parents, after an uncomplicated pregnancy. Delivery and neonatal period were normal. Birth weight 3700 g, length 54 cm and head circumference 35.8 cm. An older brother was healthy. The family history revealed nonspecific mental retardation in six male maternal relatives with neither neurological findings nor dysmorphic features.
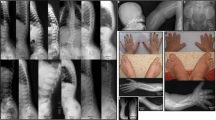
He was always a quiet baby, with somewhat delayed developmental milestones: he sat at 8 months, walked at 21 months, a few words were spoken at 3½ years. Sign language was used later. The growth followed the 50% centile. No dysmorphic features were noted apart from slight clinodactyly of fifth fingers. The neurological examination revealed moderate general hypotonia, and extensive lordosis because of hypotonia; furthermore, fine motor function was not adequate for age, showing abnormal, dystonic hand movements, corresponding to Partington syndrome (Partington sign) (Figure 1). A CT scan of the brain was normal apart from asymmetrical lateral ventricles.
39589 (c.333_334ins(GCG))
The boy was born as number 3 of three children. His mother's sister's son attended special school. Birth weight 3200 g and head circumference 34 cm. On referral, 5 years old, a small head circumference at the 10th centile was noted, but otherwise growth was normal. Language development was retarded and abnormal unconcentrated behaviour was described. Hearing and eyesight were normal. He attended special school. He had a history of epilepsy in the form of absence seizures.
41263 (c.333_334ins(GCG))
The boy was born as number 4 out of four (three healthy sisters). The parents were consanguineous and of Pakistani descent. His mother's sister's daughter showed retarded language development (no language at 5 years of age) and motor problems. Pregnancy and delivery were normal, birth weight 3700 g, length 53 cm and head circumference 35 cm. At the age of 4 years, he was described as a happy, confident child with an Angelman-like appearance with a broad mouth and low placed ears. He had general developmental delay with severe delay of language development and an ADHD-like behaviour. There was no history of seizures. He was hypotonic with affected motor function of the mouth and had difficulties chewing and swallowing. MR scanning of brain, EEG, ophthalmological and audiological investigations were normal, although delayed visual maturation was described. Investigations for Angelman syndrome, FISH and methylation analysis of SNRPN as well as mutational analysis of UBE3A, were normal.
47063 (c.333_334ins(GCG)3)
The child, aged 13 years, was born as number 2 of two. An older brother was healthy, apart from a congenital heart malformation that did not require operation. The family history revealed a maternal uncle with dyslexia and a second cousin with delayed mental and motor development. His birth weight was 3200 g. The neonatal period was normal apart from colic. The motor development was slightly delayed and he walked at 18 months old. His language development was severely delayed; he spoke only few words at the age of 3 years. Temper tantrums as well as features of the autistic spectrum were described with poor and abnormal social contact. A neurological examination revealed visual-spatial problems. An ophthalmological examination was normal. Physical assessment showed hypotonia. The developmental quotient was approximately corresponding to 50.
46609 (c.431_454del(24 bp))
A 6-year-old boy who was born as number 1 of two to consanguineous parents of Pakistani descent. A younger sister was healthy. There was no family history of mental retardation. Pregnancy and delivery were uncomplicated, birth weight unknown. There were no neonatal problems and thriving was normal in infancy. Motor development was slightly retarded, he walked at 18 months of age. At age 2 years and 11 months he spoke only few words. He was described as passive, sleeping a lot and easily distractable. At the age of 6 years he had a normal neurological examination, apart from possible problems in visual perception and possible apraxia. Physical examination showed no major dysmorphic features.
DNA extraction and mutation screening
Genomic DNA was isolated from blood leucocytes by a standard salting out method.7 Fragment size analysis of nucleotides 247–497 was carried out by PCR amplification of 50 ng of genomic DNA using fluorescent-labelled primer (6-FAM or HEX) 2563-F: 6-FAM/HEX- CCCAAGCTGCGGCGCCTGTA) and primer 2794-R: TGCGGCGCCTGGCTGATCTT) each 0.5 μ M, Failsafe premix J from Epicentre (Madison, Wisconsin, USA) in final 1 × concentration and 2.45 U Expand Long Template Enzyme Mix from Roche Applied science (Basel, Switzerland). Initial denaturation was 10 min at 95°C, followed by 26 cycles of: 94°C for 40 s, 65°C for 30 s and 68°C for 2 min. A final extension step at 68°C for 10 min ended the protocol. The fragments were analysed on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Mutational analysis of all five exons and flanking regions was performed using PCR amplification followed by direct sequencing using Big Dye Terminator v3.1 Cycle Sequencing kit from Applied Biosystems (Foster City, CA, USA) and analysis on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Protocols and primers sequences were modified from Strømme et al2 and are available upon request.
Screening for c.1347C>T
To test for the presence of c.1347C>T, exon 4 was amplified by PCR as for the mutation analysis. The PCR product was purified using QIAquick (Qiagen, Hilden, Germany) followed by digestion with restriction enzyme BanII. This will give products of the following sizes: 35, 39, 72, 191 and 244 bp for wild-type sequences and 35, 39 72 and 435 bp in case the c.1347C>T mutation is present.
Results
Screening of nucleotides 247–497 of ARX in 682 retarded males by fragment size analysis revealed four males with fragment sizes different from the control (Table 1). DNA sequencing showed the presence of the same in-frame insertion of GCG (c.333_334insGCG) in two patients (39589 and 41263), leading to the expansion of the polyalanine tract at amino acid 100–115 from 16 to 17 alanines. The mother of 39 589 was found to be a carrier of the mutation. We also investigated a cousin (mother's sister's son), who was reported to have special needs at school and a healthy brother – neither of these two boys had the mutation. The mother of 41 263 was also a carrier of the c.333_334insGCG mutation. Investigation of 188 control males with no signs of developmental delay, revealed one with the c.333_334insGCG insertion.
Patient 47 063 had an expansion of the same alanine tract by three alanines (c.333_334ins(GCG)3). Unfortunately, the mother of this patient was not available for investigation.
Patient 46 609 had a fragment smaller than the control, and DNA sequencing showed an in-frame deletion (c.431_454del(24 bp)) of the polyalanine tract at amino-acid position 144–155 from 12 to 4 alanines. The mother was a carrier of the deletion. This alteration has been reported as a probable polymorphism.1
Mutational analysis of all exons of ARX in 11 MRX families revealed one proband (29 442) with the previously described recurrent mutation c.431_454dup(24 bp) causing an expansion of the polyalanine tract at amino-acid position 144–155 from 12 to 20 alanines. Analysis of the mother and healthy brother and three other mentally retarded males from this family showed that this was a de novo mutation. The pedigree is shown in Figure 2. The fragile X syndrome was excluded in these family members.
In another MRX family, we found a proband (D301) with a silent mutation in exon 4 (c.1347C>T, G449G). This substitution was also found in his mother, his affected maternal uncle and his grandmother; however, the substitution was also found in nonaffected members of the family. This substitution has been reported previously.8 The c.1347C>T substitution was also found in one of the three MR families (proband 14 345 and his affected brother). Investigation of 89 control males with no signs of mental retardation revealed four with the c.1347C>T substitution.
Mutational analysis of all exons of ARX in 11 probands with familial autism showed no alteration of the ARX sequence.
Discussion
Mutations in ARX have recently been reported to give rise to a broad spectrum of phenotypes, from nonspecific mental retardation to syndromic forms such as Partington syndrome and XLAG. Furthermore, ARX mutations have been suggested to be more frequent in XLMR families than mutations in other known XLMR genes, apart from FMR1. We wanted to investigate whether a mutation in ARX is a common cause of mental retardation in males in Denmark.
Two recurrent mutations have been reported, c.333_334ins(GCG)7 and c.431_454dup(24 bp), leading to the expansion of polyalanine tracts at amino-acid positions 100–115 and 144–155, respectively.1, 2 Screening for size alterations in the region from nucleotide 247 to 497 in samples from 682 males revealed only four patients with altered ARX sequence. One patient had a previously described deletion, and in two cases the sequence alteration lead to expansion of a polyalanine tract (amino acid 100–155) by one alanine, whereas the fourth case represents expansion of the same tract by three alanines. The expansion of this polyalanine tract with the addition of either two or seven alanines was previously reported in one MRX family and two ISSX families, respectively.1, 9 The addition of one alanine to the tract was found in one out of 188 control samples, indicating that this is a polymorphism. Analysis of family members of 39 589 (one of the two cases with the addition of one alanine, c.333_334insGCG) revealed that a cousin with special needs at school did not have the mutation, supporting that this is a rare polymorphism. Unfortunately, it was not possible to study family members of the other families. It is thus unresolved if the addition of three alanines (c.333_334ins(GCG)3) may be disease causing.
In comparison, we found 15 males with fragile X syndrome in the same group of patients (these were excluded from the group prior to screening the ARX gene). Thus, the recurrent mutations in ARX seem to be more rare than the fragile X syndrome, at least in this group of patients.
At least seven other known genes harbour alanine tracts that have been shown to expand and cause disease in human. These include six transcription factors: ZIC2 causing holoprosencephaly, HPE (from 15 to 25 alanines),10 HOXA13 causing hand–foot–genital syndrome, HFGS (from 18 to 24, 26 alanines),11 FOXL2 causing blepharophimosis/ptosis/epicantus inversus syndrome (from 14 to 24 alanines),12 RUNX2 (CBFA1) causing cleidocranial dysplasia, CCD (from 17 to 27 alanines),13 HOXD13 causing synpolydactyly, SPD (from 15 to 22–25, 29 alanines),14 PHOX2B causing congenital central hypoventilation syndrome, CCHS (from 20 to 25, 26, 27, 28, 29 alanines)15 and SOX3 causing XLMR and growth hormone deficiency, GHD (from 15 to 26 alanines).16 Expansion sizes vary from 5 to 14 extra alanines. One nontranscription factor is known: PABPN1 where expansion from 6 to 7–13 alanines causes oculopharyngeal muscular dystrophy, OPMD.17 All these genes exhibit an autosomal dominant inheritance; however, expansion by one alanine in the PABPN1 gene causes a recessive form of OPMD or act as a modifier of a dominant phenotype. However, PABPN1 is not a transcription factor in contrast to ARX. The smallest disease-causing expansion in a transcription factor is of five alanines from 20 to 25 (PHOX2B), and the smallest disease-causing allele in transcription factors has 20 alanines (ARX). Taken together, it seems that c.333_334insGCG that causes an expansion from 16 to 17 alanines is a rare polymorphism, whereas further studies are necessary to clarify the significance of c.333_334ins(GCG)3 (expansion from 16 to 19 alanines).
We did not find mutations in ARX in 11 families with familial autism; however, the material is too small to draw any conclusions about the implication of ARX in autism, given the genetic heterogeneity of this condition.
Our data concerning the c.1347C>T substitution indicate that this is a polymorphism. However, in silico analysis of the altered sequence using the ESEfinder (http://exon.cshl.edu/ESE/)18 shows that c.1347C>T creates a strong binding site for SRp55. SR proteins are splicing factors binding to exonic splice enhancers.19 Thus, it could be speculated that c.1347C>T could act as a modifier gene.
It is notable that we only found the recurrent mutation c.431_454dup(24 bp) once among the MRX families (which in fact turned out to be a de novo case).
We conclude that the previously reported polyalanine expansions in ARX are probably not a frequent cause of nonspecific mental retardation, and screening for the mutations is not warranted at the moment as part of the assessment of a child with global developmental delay without a family history.
References
Bienvenu T, Poirier K, Friocourt G et al: ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet 2002; 11: 981–991.
Stromme P, Mangelsdorf ME, Shaw MA et al: Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 2002; 30: 441–445.
Kitamura K, Yanazawa M, Sugiyama N et al: Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet 2002; 32: 359–369.
Turner G, Partington M, Kerr B, Mangelsdorf M, Gecz J : Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am J Med Genet 2002; 112: 405–411.
Ropers HH, Hoeltzenbein M, Kalscheuer V et al: Nonsyndromic X-linked mental retardation: where are the missing mutations? Trends Genet 2003; 19: 316–320.
Miura H, Yanazawa M, Kato K, Kitamura K : Expression of a novel Aristaless related homeobox gene ‘Arx’ in the vertebrate telencephalon, diencephalon and floor plate. Mech Dev 1997; 65: 99–109.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Kato M, Das S, Petras K et al: Mutations of ARX are associated with striking pleiotropy and consistent genotype–phenotype correlation. Hum Mutat 2004; 23: 147–159.
Stromme P, Mangelsdorf ME, Scheffer IE, Gecz J : Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev 2002; 24: 266–268.
Brown LY, Odent S, David V et al: Holoprosencephaly due to mutations in ZIC2: alanine tract expansion mutations may be caused by parental somatic recombination. Hum Mol Genet 2001; 10: 791–796.
Goodman FR, Bacchelli C, Brady AF et al: Novel HOXA13 mutations and the phenotypic spectrum of hand–foot–genital syndrome. Am J Hum Genet 2000; 67: 197–202.
Crisponi L, Deiana M, Loi A et al: The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 2001; 27: 159–166.
Mundlos S, Otto F, Mundlos C et al: Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997; 89: 773–779.
Muragaki Y, Mundlos S, Upton J, Olsen BR : Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 1996; 272: 548–551.
Amiel J, Laudier B, Attie-Bitach T et al: Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat Genet 2003; 33: 459–461.
Laumonnier F, Ronce N, Hamel BC et al: Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet 2002; 71: 1450–1455.
Brais B, Bouchard JP, Xie YG et al: Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 1998; 18: 164–167.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR : ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003; 31: 3568–3571.
Cartegni L, Chew SL, Krainer AR : Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 2002; 3: 285–298.
Acknowledgements
We thank Annie Sand for technical assistance. We thank Jette Bune Rasmussen for making the figures. We thank Professor Jozef Gécz for helpful communication concerning the PCR procedure.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grønskov, K., Hjalgrim, H., Nielsen, IM. et al. Screening of the ARX gene in 682 retarded males. Eur J Hum Genet 12, 701–705 (2004). https://doi.org/10.1038/sj.ejhg.5201222
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201222
Keywords
This article is cited by
-
Screening of the duplication 24 pb of ARX gene in Moroccan patients with X-linked Intellectual Disability
BMC Research Notes (2021)
-
Genetics, Molecular Biology, and Phenotypes of X-Linked Epilepsy
Molecular Neurobiology (2014)
-
MRX87 family with Aristaless Xdup24bp mutation and implication for polyAlanine expansions
BMC Medical Genetics (2007)
-
Mutation screening of brain-expressed X-chromosomal miRNA genes in 464 patients with nonsyndromic X-linked mental retardation
European Journal of Human Genetics (2007)
-
Mutational screening of ARX gene in Brazilian males with mental retardation of unknown etiology
Journal of Human Genetics (2006)