Abstract
The 3′-untranslated region (3′UTR) of some vertebrate dystrophin genes shows an extraordinary degree and extent of conservation (better than that of many coding regions), a phenomenon that remains unexplained. We examine novel sequence and mutational data to explore the possible reasons for this. We show that loss of the human dystrophin 3′UTR is sufficient to cause Becker muscular dystrophy with pronounced reduction in dystrophin protein levels. The acquisition of dystrophin 3′UTR sequence from an amphibian and a cartilaginous fish allows us to refine previously identified functionally constrained regions which might account for the observed phenotype. These comprise (a) the open reading frame encoding the ancestral ‘alternative’ amphipathic C-terminal α-helix, normally removed from adult dystrophin by inclusion of a poorly conserved frameshifting penultimate exon, and (b) two highly conserved untranslated regions (‘Lemaire A’, 350 nucleotides and ‘Lemaire D’, 250 nucleotides) separated by a non-conserved 700–2000-nucleotide spacer. We consider the possibility that the 3′UTR may represent a significant target for pathogenic mutations.
Similar content being viewed by others
Introduction
The 3′-untranslated regions (3′UTRs) of eukaryotic mRNAs are often dismissed as incidental products of transcription, filling in between the sites of translation termination and transcript polyadenylation. This impression is reinforced by the low level of conservation of some 3′UTRs, a fact exploited by their specific use as hybridisation probes to avoid unwanted cross-reactivity which might ensue from the use of (better conserved) coding regions.
Only in a minority of cases has a function been ascribed to specific elements residing within 3′UTRs. These generally entail interaction between RNA-binding proteins and short sequences and/or secondary structure motifs in the 3′UTR. Effects mediated by such interactions include regulation of transcript stability, specification of subcellular transcript localisation, regulation of translation and the incorporation of selenocysteine.1,2
Empirically, however, it has been noted3 that a substantial proportion of vertebrate genes have 3′UTRs which contain regions of several hundred nucleotides whose degree of conservation approaches or even exceeds that of their respective coding region. This conservation through evolution seems to imply some form of strong and physically extended functional constraint on sequence; no currently known biological function appears to be able to apply such a constraint.
The dystrophin gene, mutations in which cause the human genetic disorders Duchenne and Becker muscular dystrophy (DMD, BMD), was noted to have an unusually well conserved 3′UTR when the chicken orthologue was first described.4 The human, mouse and chicken dystrophin 3′UTRs are large (2400–2700 nucleotides), with regions of high conservation at each ends, separated by a region of relatively low conservation. The significance of these regions both for the function of the dystrophin gene and as targets for pathogenic mutagenesis is currently unknown.
In this report we extend studies of evolutionary conservation of the 3′UTR across vertebrate genes for dystrophin and related proteins. We find that regions at both ends of the dystrophin 3′UTR have been strongly conserved for >420 My, and explore potential reasons for this. We also refine the characterisation of a dystrophin gene mutation which results in the loss of the wild-type 3′UTR from the transcript, and report the consequences for the protein and phenotype.
Materials and Methods
3′-RACE with vectorette
3′ rapid amplification of cDNA ends (3′-RACE) with vectorette was performed as follows. Five hundred ng of total RNA extracted from skeletal muscle of Xenopus laevis or Scyliorhinus canicula were reverse transcribed using MMLV reverse transcriptase (Roche), using the primer 5′-tgaacgtcccgggaaacagtccggagacgtgcagtttttttttttttvnn-3′ (IUPAC single letter code). This was then amplified by polymerase chain reaction (PCR) using the adaptor primer 5′-tgaacgtcccgggaaacagtccggagacgtg-3′ together with primers complementary to our previously published X. laevis and S. canicula dystrophin coding sequences (X99700, X99702, respectively; sequences available on request). The resulting 3′-RACE products were purified, digested with XmaI, and ligated to a Y-shaped vectorette (formed by annealing of the oligonucleotides VecC, 5′-cgaatcgtaagcggccgcagacgacgatctgtcctctcctt-3′ and VecZ-X, 5′-ccggaaggagaggacgctgtctgtcg-3′). These were then re-amplified using a nested dystrophin-specific primer plus vectorette primer 224C (5′-cgaatcgtaagcggccgcagacgacgatct-3′) and sequenced.
Bioinformatics
Sequence alignments were carried out using CLUSTAL within Vector NTI Suite 6.0 (InforMax). Untranslated sequences were examined for potential secondary structure using RNAstructure 3.65 and for known 3′UTR functional elements using UTRScan.1
Clinical description of patients
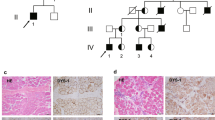
The three brothers 4482–4484, currently aged 19, 16 and 14 years, respectively, suffer from a syndrome of progressive skeletal myopathy, GK deficiency and adrenal hypoplasia congenita, with undescended testes and moderate learning difficulties. The possible aetiology of their learning difficulties has been discussed elsewhere.6 The oldest brother (4482) was referred at the age of 4 years with difficulty in running and climbing stairs. The progression of his myopathy has since been relatively slow, and in his late teens he can still hop and jump, can walk for 400 m without tiring, and can rise from the floor in 4 s using Gower's sign. He has mild calf hypertrophy and Achilles tendon contractures. The second brother (4483), although younger, is more severely affected in terms of his myopathy (tiring after walking 10 m), endocrinology and cognitive problems. The youngest brother (4484) can still jump, hop and run and walks unlimited distances. The phenotype of all three is clearly that of Becker, rather than Duchenne, muscular dystrophy.
Localisation of deletion breakpoint
We had previously shown that the exon 78 of the dystrophin gene and exon 8 of the IL1RAPL1 gene were spared in patients 4482–4484, while exon 79 of the dystrophin gene and exons 9–11 of the IL1RAPL1 gene were at least partially deleted. We used the sequence of the dystrophin intron 78 (4.8 kb; accession number AC023414) and partial fragmentary sequence of IL1RAPL1 intron 8 (>20 kb; accession number AC005748) to design sequence-tagged sites (sequences available on request), which were used in an iterative process, each round serving to halve the region of the intron known to harbour the deletion breakpoint. After coarsely delineating the positions of the breakpoints to the order of 1–2 kb, primers from the furthest retained reactions in the two genes were combined and PCR across the breakpoint was attempted. The product was sequenced on an ABI Prism 377 DNA Sequencer using dRhodamine dye terminators. At this time it was also necessary to close a gap in the wild-type IL1RAPL1 intron 8 sequence by analysing a PCR product from a control individual (accession number AF375549).
Histology and immunofluorescence
A needle biopsy from the quadriceps of one brother was taken at age 7 years and was rapidly frozen in isopentane cooled in liquid nitrogen. Cryostat sections were stained with histological and histochemical techniques. Unfixed cryostat sections were immunolabelled with a panel of antibodies to the rod (Dys3) and C terminal domain (Dys2) of dystrophin. These were visualised with a biotinylated-streptavidin-Texas Red method and examined by epifluorescence with a Leica Aristoplan microscope.7
Results
Sequence conservation of the dystrophin 3′UTR
The unusually high degree of conservation of parts of the dystrophin 3′UTR has been noted previously.4 Lemaire and colleagues described three regions of high similarity (80–90% identity) between the 3′UTRs of human and chicken dystrophin mRNAs. These regions (herein referred to as Lemaire A, C and D) are distributed along the length of the 3′UTRs, interspersed with regions of no detectable homology.
We extended this observation by isolating the corresponding sequences from the amphibian X. laevis and the cartilaginous fish S. canicula, using 3′RACE from our previously characterised dystrophin coding sequences8 (accession numbers X99700, X99702). This yielded 466 nt of 3′UTR for Xenopus and 1534 nt for Scyliorhinus (accession numbers AF375546 and AF375547, respectively).
The unexpectedly short Xenopus sequence may be incomplete; although the apparent polyA tail is preceded by two potential polyadenylation signals absent from the other species, the 3′RACE may have primed from a genomically encoded run of A residues. The Xenopus sequence shows high continuous similarity to the Lemaire A region of human and chicken dystrophin 3′UTRs.
The dogfish sequence, although less than two-thirds of the length of its human counterpart, contains homologous sequences at both ends (Lemaire A and D) and uses a polyadenylation signal in an almost identical context to the human transcript. We believe this 3′UTR sequence to be complete.
A dotplot of the entire human and dogfish 3′UTRs reveals only two regions of significant homology over a wide range of window size and stringency (Figure 1A). The first region, which corresponds to the Δ78 open reading frame (see below) and ‘Lemaire A’, extends over the first ∼400 nucleotides of the 3′UTR. The second, which corresponds approximately to ‘Lemaire D’, comprises the last ∼250 nucleotides of the 3′UTR. The intervening region (1966 bp in human, 768 bp in dogfish) shows no significant conservation, with no vestige of the ‘Lemaire C’ region previously described in chicken.4
Conservation of the dystrophin 3′UTR. (A) Dotplot of the human and dogfish 3′UTRs, from the muscle transcript stop codon to the polyadenylation site. Boxes indicate regions of high homology for which alignments are shown in Figures 1B, 2A and 2B. (B) Alignment of notional translation of the Δ78 open-reading frame from selected species. Black and grey highlights indicate identity and similarity to human sequence, respectively. (C) Helical wheel projection of the 23 residues from the Δ78 ORF predicted to form an amphipathic α-helix. Shaded residues are non-polar.
Δ78 transcripts; conservation at the amino acid level
An explanation of the conservation of the first ∼100 nucleotides of the dystrophin 3′UTR is straightforward. In a subset of human and mouse dystrophin transcripts the penultimate exon (exon 78) has been found to be omitted, resulting in a reading frame shift which brings into register a novel open reading frame (ORF) in exon 799,10 – this results in a substitution of the ‘normal’ (muscle-type) C-terminal 14 amino acids with 32 new ones (to which we refer as the Δ78 ORF). The proportion of Δ78 transcripts is much higher in embryonic brain and muscle, falling dramatically as development progresses.10 We have previously noted the conservation of this shifted ORF in Xenopus and dogfish.8 The recent description of C-terminal dystrophin sequence from invertebrates such as sea urchin,11 nematode12 and (to a lesser extent) fruitfly13 shows the Δ78 sequence to be the ancestral one, retained in embryonic dystrophin (Figure 1B), but not in the adult muscle isoform or the two paralogous proteins utrophin and DRP2.
The function of the Δ78 peptide is not clear; it may represent a site of interaction for a protein which is only required in early stages of development. The amino acid content is highly acidic (22%) and hydrophobic (30%), and is strongly predicted to form an α-helix (using the neural net secondary structure prediction program PHDsec14). Helical wheel projection of all but the last few amino acids reveals the potential to form a strongly amphipathic 23-residue helix (VGS…–VMT), with one side hydrophobic and the other almost entirely charged or polar (Figure 1C). This potential to form an amphipathic helix is conserved in all of the above mentioned sequences except Drosophila.
It is striking that the sequence conservation of exon 78 is itself rather poor (zebrafish exon 78 is barely recognisable and even contains a stop codon15,16), and we suggest that its purpose may indeed be solely to afford a means of removing the amphipathic Δ78 peptide from adult dystrophin.
Conservation at the nucleotide level
Duret and co-workers3 define Highly Conserved Regions (HCRs) as non-coding regions in excess of 100 nt which exhibit >70% identity between species which diverged >300 My ago. They reason that sequences not subject to selective pressure will accumulate sufficient mutations over 300 My to reduce their similarity to ∼30%, ie approximately that of unrelated sequences. We find the region which spans 400 nt 3′ of the dystrophin Δ78 stop codon (ie Lemaire A) to be 73% identical between human and dogfish, species that diverged >420 My ago, an achievement which clearly marks it as an HCR. The HCR is more highly conserved between human and mouse (97%) than is the coding region (93%; see Table 1). We consider the HCR to end at the point indicated in Figure 2A because (a) the human/mouse identity drops to 68%, which is similar to the 71% mean identity found between 2820 orthologous human and mouse 3′UTRs,17 (b) the Xenopus transcript appears to terminate and (c) the similarity between human and dogfish drops below significance (no similarity is seen even on a dotplot, a method which makes no assumptions about position or orientation).
The Lemaire A and Lemaire D HCRs. (A) Alignment of nucleotide sequences starting immediately after the Δ78 stop codon and continuing until homology between human and dogfish drops below significance – the ‘Lemaire A’ HCR. (B) Alignment of the last ∼300 nucleotides of dystrophin 3′UTRs – the ‘Lemaire D’ HCR. Key: black and grey highlight, conserved in all and all-but-one species respectively; underline, potential polyadenylation signals; dashes, gaps introduced by CLUSTAL to maximise alignment. Accession numbers: M18533, M68859, X13369, AF375546, AF375547, BE120428, BE750440, BF198600, AI723149, AJ012469, AF277386, AF304204. Bold boxes in A and B indicate predicted conserved stem-loop structures. Light box indicates region previously implicated in vigilin binding.20
We ascertained whether similar levels of conservation are found in the proximal 3′UTRs of other members of the dystrophin family. Table 1 shows the percentage similarity between aligned sequences of human and mouse transcripts of dystrophin, utrophin and DRP2 (using the last 400 nt of the coding region and the first 800 nt of the 3′UTR). The percentage identity over each indicated block is given. The coding region conservation for all three paralogues is similar (89–93%), and modestly higher than the 85% human/mouse average17 (bottom line of Table 1). The DRP2 transcript (which, like dystrophin, is encoded by an X-linked gene18) shows average human/mouse similarity in its 3′UTR, indicating a lack of selective pressure on this region. The mouse and human utrophin 3′UTRs show a degree of conservation intermediate between that of DRP2 and dystrophin, which drops to unselected levels after 400 nt. This region has recently been implicated in the targeting and stability of utrophin transcripts.19 The dystrophin, utrophin and DRP2 3′UTRs do not show significant similarity to each other. This maintenance of HCRs between orthologues but not between paralogues has been previously noted.3
The distal HCR, ‘Lemaire D’, also shows a high level of identity between human and dogfish, with 80% identity over the 3′ 140 nt, including the polyadenylation signal and a region previously implicated in vigilin binding.20 It seems safe to assume that no selective pressure is acting on the intervening ‘spacer’ sequence, although it may have certain size constraints.
Consequences of loss of the dystrophin 3′UTR
A large deletion has been described21 which removes part of the dystrophin gene, together with the glycerol kinase and DAX1 genes, resulting in a contiguous gene deletion syndrome in three brothers (patients 4482–4484). The 1.8-Mb deletion6 also removed the MAGEB gene cluster,22 the gene for the testis-specific ferritin heavy chain FTHL17,23 and part of a gene encoding a novel member of the interleukin-1/interleukin-18 receptor family, IL1RAPL1.6
We here set out to characterise the proximal breakpoint of the deletion in order to explore the aetiology of the muscular dystrophy in these patients. The breakpoints were localised as described in Materials and methods (Figure 3A), and amplification across the deletion junction was performed in patient 4482. Comparison of the resulting chimeric genomic sequence (accession number AF375548) with that of the newly resolved region of IL1RAPL1 intron 8 (AF375549) and dystrophin intron 78 (AC023414) clearly revealed a clean deletion breakpoint (Figure 3B). This is very close to the middle of the 21.5-kb IL1RAPL1 intron, but only 455 bp 3′ of exon 78 of the dystrophin gene.
The deletion breakpoint in family 4482. (A) Partial maps of the wild-type dystrophin and IL1RAPL1 genes, drawn to scale from genomic sequence, together with the deduced map of affected members of family 4482. Xpter is to the right in each case. Exon numbering is from Carrié et al. who showed that IL1RAPL1 has 11, rather than 10, exons.32 Heavy dashed line links deletion breakpoints. (B) Sequences of control individuals and family 4482 patients around the deletion breakpoint.
We have previously shown6 that patient 4482′s dystrophin transcript carries a novel 3′ exon in the place of the wild-type exon 79; this arises via the use of a cryptic splice acceptor site on the antisense strand of intron 6 of the fused IL1RAPL1 gene (‘4482UTR’; accession number AF181286). This work also showed (by 3′RACE from dystrophin exon 76) that the proximity of the breakpoint to exon 78 (455 bp; see above) does not affect the splicing of this exon in muscle transcripts. Thus the only discernible qualitative effect of the mutation on the dystrophin transcripts is the substitution of the last exon. Although insufficient RNA was available to perform a quantitative mRNA assay, the availability of muscle biopsy material enabled an assessment of the consequences of the mutation at the protein level.
The muscle biopsy from one brother showed dystrophic features with variation in fibre size, whorled fibres, evidence of regeneration and excess endomysial connective tissue in some areas (Figure 4A). Type 1 fibres were predominant. Immunolabelling for dystrophin showed a pronounced reduction with only slight traces detectable with antibodies to both rod domain (Figure 4B) and C terminus of dystrophin. β-dystroglycan was also reduced (data not shown). Although the epitope for the C-terminal antibody (the last 17 amino acids) is expected to be severely disrupted by the mutation, the signal from the rod domain antibody (Figure 4B) should be an accurate reflection of the amount of dystrophin present.
Discussion
The 3′UTR of the dystrophin transcript contains two regions which qualify as HCRs as defined3 by Duret et al In this work we have extended previous studies of these sequences by examining them in an amphibian and a cartilaginous fish, and explored the significance of their conservation. The function of HCRs is very poorly understood; indeed, the continuous high degree of conservation over hundreds of bases is hard to reconcile with our current concepts of the short target sequences of RNA-binding proteins. The only specific role proposed for any portion of the dystrophin HCRs is the binding of the ubiquitous KH domain protein vigilin, which has been found to interact with part of the ‘Lemaire D’ HCR in vitro.20 However the proposed vigilin binding site (see Figure 2B) is far from being the most highly conserved part of the HCR, and the biological significance of this interaction remains unclear. At a more empirical level, preliminary work24 suggests that addition of part of the native 3′UTR to dystrophin expression constructs increases expression following germ line gene transfer in vivo and transient transfection in vitro.
Mechanisms other than sequence-specific binding proteins might better account for the extended conservation of HCRs. These might involve functionally important RNA hybridisation, either intramolecular (in the form of secondary structure) or intermolecular (base-pairing with another RNA species). We tested for the likelihood of conserved secondary structure by using RNAstructure v3.65 to calculate minimum energy structure predictions for human and dogfish Lemaire A and D sequences. Although both yielded robust secondary structures which varied little between optimal models and were largely insensitive as to whether the Lemaire A and D regions were folded in isolation or as part of a single larger RNA molecule, few isolated elements (two stem-loops indicated in Figure 2A and B) were conserved between the two organisms. We therefore think it unlikely that secondary structure is a prime determinant of the observed sequence conservation. A further possibility is that RNA molecule(s) encoded by an independent genomic locus bind to sequences within Lemaire A and D; the need for the two RNAs to maintain pairing would constrain their primary sequences. A precedent for this is the let-7 system, first decribed in Caenorhabditis elegans,25 but since found to be conserved throughout the animal kingdom.26 The let-7 locus encodes a non-coding transcript which is processed to form a 21 nt RNA molecule.26 This hybridises to conserved elements in the 3′UTRs of a range of developmentally regulated transcripts,25 effecting temporal control of their translation. In order to test the possibility that such a mechanism might explain the conservation of Lemaire A and D, we searched human genomic and expressed sequence databases for region which might form RNA–RNA hybrids with the dystrophin 3′UTR, taking into account the possibility of G-U pairing. Although some purine-rich regions showed some possibility of weak pairing, these were all of a rather simple sequence, showing none of the complexity of the HCRs.
DMD and BMD are common monogenic disorders which range in severity from moderately disabling (mild BMD) to highly progressive and ultimately lethal (classic DMD). This has led to substantial mutational analysis and thousands of independent mutations have been described. Despite this, and the apparent functional importance of the 3′UTR implied by its extraordinary degree of conservation, very few mutations involving the 3′UTR of the dystrophin gene have been described, a fact that might be entirely explained by the pragmatic decision not to include this region in most screening protocols.
A number of large dystrophin gene deletions have been reported which remove multiple 3′ exons, including exon 79 (and sometimes neighbouring genes). It is clear from the instance of a BMD patient lacking exons 73–7927 that loss of the entire 3′UTR is compatible with a fairly mild muscle phenotype. We note, however, that one patient has been described28 in whom a 13-nucleotide deletion in the Δ78 coding region is expected to lead to substitution of the C-terminal 21 Δ78 codons with five new ones (DDLGRAMESLVSVMTDEEGAE* → ERWSP*). This patient has a DMD phenotype and no other reported mutation; the pathogenic relevance of this deletion remains unclear. No variants of the HCRs have been reported, and only one silent change in the Δ78 ORF has been described.29
In this paper we have described the precise nature of a mutation which results in the specific loss of exon 79 from the dystrophin gene. As a consequence, exon 78 is spliced directly onto a cryptic exon on the antisense strand of an intron of the IL1RAPL1 gene.6 We show here that this results in a substantial reduction in the level of dystrophin protein in skeletal muscle. A number of potential mechanisms for this reduction can be adduced a priori: (a) the poorly conserved C-terminal three amino acids of the muscle-type dystrophin (−DTM) are replaced by six new ones (−ALCCHT); (b) the highly conserved embryonic Δ78 ORF, encoding a potential amphipathic α-helix, is lost; (c) the cryptic final exon used by the dystrophin transcript in these patients may lead to inefficient splicing and/or polyadenylation, thereby reducing mRNA levels; (d) nonsense-mediated decay may occur if a splicing event occurs within the novel 3′UTR; (e) one or both of the HCRs in the normal 3′UTR may be needed for proper termination, processing, nuclear export, stability, translation or subcellular localisation of the dystrophin transcript. The absence of the HCRs may therefore result in low levels of a fully functional dystrophin protein; we have previously shown30 that such a solely quantitative defect is sufficient to cause BMD.
Given their high level of conservation, it seems likely that point mutations affecting Lemaire A or D might result in a BMD phenotype; examination of these parts of the 3′UTR should therefore be considered for mutation screening in the case of BMD patients without mutations which affect the coding sequence. As deletion of the entire 3′UTR only gives a mild phenotype, a severe DMD phenotype is unlikely to result from loss-of-function 3′UTR mutations. It is possible, however, that mutations in the 3′UTR which create novel splice donor sites might cause a severe phenotype by activating nonsense-mediated decay. As a number of recently reported dystrophin gene mutation screening systems29,31 have begun to incorporate exon 79, some idea of the contribution of the HCRs to BMD pathogenesis may be forthcoming.
Most initiatives aimed at gene therapy of the dystrophinopathies currently use the dystrophin coding region in an entirely heterologous sequence context, largely owing to vector size constraints. The phenotype of our patients suggests that the use of a heterologous 3′UTR may compromise dystrophin transcript function. Ultimately an understanding of the function of these highly conserved elements, whether from the identification of more subtle 3′UTR pathogenic mutations or from in vivo and in vitro experimentation, will enable the rational design of a ‘mini-3′UTR’, which may prove essential for proper dystrophin expression.
References
Pesole G, Grillo G, Larizza A, Liuni S . The untranslated regions of eukaryotic mRNAs: structure, function, evolution and bioinformatic tools for their analysis Brief Bioinform 2000 1: 236–249
Hazelrigg T . The destinies and destinations of RNAs Cell 1998 95: 451–460
Duret L, Dorkeld F, Gautier C . Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression Nucl Acids Res 1993 21: 2315–2322
Lemaire C, Heilig R, Mandel JL . The chicken dystrophin cDNA: striking conservation of the C-terminal coding and 3′ untranslated regions between man and chicken EMBO J 1988 7: 4157–4162
Mathews DH, Sabina J, Zuker M, Turner DH . Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure J Mol Biol 1999 288: 911–940
Jin H, Gardner RJ, Viswesvaraiah R, Muntoni F, Roberts RG . Two novel members of the interleukin-1 receptor gene family, one deleted in Xp22.1-Xp21.3 mental retardation Eur J Hum Genet 2000 8: 87–94
Sewry CA, Lu Q . Immunological reagents and amplification systems in Bushby K, Anderson LVB (eds) Muscular dystrophy: methods and protocols Ottowa: Humana Press 2001 pp 325–338
Roberts RG, Bobrow M . Dystrophins in vertebrates and invertebrates Hum Mol Genet 1998 7: 589–595
Feener CA, Koenig M, Kunkel LM . Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus Nature 1989 338: 509–511
Bies RD, Phelps SF, Cortez MD, Roberts R, Caskey CT, Chamberlain JS . Human and murine dystrophin mRNA transcripts are differentially expressed during skeletal muscle, heart, and brain development Nucl Acids Res 1992 20: 1725–1731
Wang J, Pansky A, Venuti JM, Yaffe D, Nudel U . A sea urchin gene encoding dystrophin-related proteins Hum Mol Genet 1998 7: 581–588
Bessou C, Giugia JB, Franks CJ, Holden-Dye L, Segalat L . Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission Neurogenetics 1998 2: 61–72
Greener MJ, Roberts RG . Conservation of components of the dystrophin complex in drosophila(1) [In Process Citation] FEBS Lett 2000 482: 13–18
Rost B, Sander C . Improved prediction of protein secondary structure by use of sequence profiles and neural networks Proc Natl Acad Sci USA 1993 90: 7558–7562
Chambers SP, Dodd A, Overall R et al. Dystrophin in adult zebrafish muscle Biochem Biophys Res Commun 2001 286: 478–483
Bolanos-Jimenez F, Bordais A, Behra M et al. Molecular cloning and characterization of dystrophin and Dp71, two products of the Duchenne Muscular Dystrophy gene, in zebrafish Gene 2001 274: 217–226
Makalowski W, Boguski MS . Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences Proc Natl Acad Sci USA 1998 95: 9407–9412
Roberts RG, Freeman TC, Kendall E et al. Characterization of DRP2, a novel human dystrophin homologue Nat Genet 1996 13: 223–226
Gramolini AO, Belanger G, Jasmin BJ . Distinct regions in the 3′ untranslated region are responsible for targeting and stabilizing utrophin transcripts in skeletal muscle cells J Cell Biol 2001 154: 1173–1183
Kanamori H, Dodson RE, Shapiro DJ . In vitro genetic analysis of the RNA binding site of vigilin, a multi- KH-domain protein Mol Cell Biol 1998 18: 3991–4003
Love DR, Flint TJ, Marsden RF et al. Characterization of deletions in the dystrophin gene giving mild phenotypes Am J Med Genet 1990 37: 136–142
Lurquin C, De Smet C, Brasseur F et al. Two members of the human MAGEB gene family located in Xp21.3 are expressed in tumors of various histological origins Genomics 1997 46: 397–408
Wang PJ, McCarrey JR, Yang F, Page DC . An abundance of X-linked genes expressed in spermatogonia Nat Genet 2001 27: 422–426
Wells KE . PhD Thesis University of London 2000
Reinhart BJ, Slack FJ, Basson M et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans Nature 2000 403: 901–906
Pasquinelli AE, Reinhart BJ, Slack F et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA Nature 2000 408: 86–89
McCabe ER, Towbin J, Chamberlain J et al. Complementary DNA probes for the Duchenne muscular dystrophy locus demonstrate a previously undetectable deletion in a patient with dystrophic myopathy, glycerol kinase deficiency, and congenital adrenal hypoplasia J Clin Invest 1989 83: 95–99
Nigro V, Nigro G, Esposito MG et al. Novel small mutations along the DMD/BMD gene associated with different phenotypes Hum Mol Genet 1994 3: 1907–1908
Mendell JR, Buzin CH, Feng J et al. Diagnosis of Duchenne dystrophy by enhanced detection of small mutations Neurology 2001 57: 645–650
Roberts RG, Passos-Bueno MR, Bobrow M, Vainzof M, Zatz M . Point mutation in a Becker muscular dystrophy patient Hum Mol Genet 1993 2: 75–77
Bennett RR, Dunnen J, O'Brien KF, Darras BT, Kunkel LM . Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing BMC Genet 2001 2: 17
Carrie A, Jun L, Bienvenu T et al. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation Nat Genet 1999 23: 25–31
Acknowledgements
We are grateful to the Medical Research Council, the Muscular Dystrophy Campaign and the Generation Trust for funding this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Greener, M., Sewry, C., Muntoni, F. et al. The 3′-untranslated region of the dystrophin gene – conservation and consequences of loss. Eur J Hum Genet 10, 413–420 (2002). https://doi.org/10.1038/sj.ejhg.5200822
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200822
Keywords
This article is cited by
-
A microRNA binding site polymorphism in the 3′ UTR region of VEGF-A gene modifies colorectal cancer risk based on ethnicity: a meta-analysis
Journal of the Egyptian National Cancer Institute (2022)
-
Are CSNK2A1 gene mutations associated with retinal dystrophy? Report of a patient carrier of a novel de novo splice site mutation
Journal of Human Genetics (2018)
-
Abnormal splicing switch of DMD’s penultimate exon compromises muscle fibre maintenance in myotonic dystrophy
Nature Communications (2015)
-
A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies
BMC Genomics (2008)
-
The dystrotelin, dystrophin and dystrobrevin superfamily: new paralogues and old isoforms
BMC Genomics (2007)