Abstract
Six breakpoint regions for rearrangements of human chromosome 15q11-q14 have been described. These rearrangements involve deletions found in approximately 70% of Prader-Willi or Angelman's syndrome patients (PWS, AS), duplications detected in some cases of autism, triplications and inverted duplications. HERC2-containing (HEct domain and RCc1 domain protein 2) segmental duplications or duplicons are present at two of these breakpoints (BP2 and BP3) mainly associated with deletions. We show here that clusters containing several copies of the human chromosome 15 low-copy repeat (LCR15) duplicon are located at each of the six described 15q11-q14 BPs. In addition, our results suggest the existence of breakpoints for large 15q11-q13 deletions in a proximal duplicon-containing clone. The study reveals that HERC2-containing duplicons (estimated on 50–400 kb) and LCR15 duplicons (∼15 kb on 15q11-q14) share the golgin-like protein (GLP) genomic sequence. Through the analysis of a human BAC library and public databases we have identified 36 LCR15 related sequences in the human genome, most (27) mapping to chromosome 15q and being transcribed. LCR15 analysis in non-human primates and age-sequence divergences support a recent origin of this family of segmental duplications through human speciation.
Similar content being viewed by others
Introduction
Human chromosome 15q11-q14 is rearranged in several disorders and structural abnormalities. These include de novo deletions accounting for approximately 70% of cases of Prader-Willi (PWS; [MIM 176270]) and Angelman syndromes (AS; [MIM 105830]);1 ‘inverted duplications’, known as inv dup(15) or idic(15);2 and interstitial duplications, some present in cases of autism,3,4,5 or triplications.6,7,8,9 Some of the 15q11-q14 rearrangements are relatively frequent, with deletions on approximately 1/10 000 of live births and inv dup(15) in approximately 2/10 000 of live births.1,2 Amos-Landgraf et al (1999)10 and Christian et al (1999)11 have shown that relatively large duplicon structures (50–400 kb) map to the breakpoint regions at 15q11 and 15q13. The hypothesis that 15q proximal rearrangements were due to repeated DNA sequences was first proposed by Donlon et al (1986),12 based on the identification of multiple copies of certain STSs. The 15q11-q13 duplicons identified by Amos-Landgraf et al (1999)10 and Christian et al (1999)11 are associated to at least two of the previous described breakpoint regions, one proximal (BP2) and the other distal (BP3) (Figure 1). These duplicons contain at least seven different expressed sequences and are thought to have been generated approximately 20 million years ago. One of the duplicon-containing expressed sequences is the HERC2 gene (HEct domain and RCc1 domain protein 2; also known as ERY-1),13 which has been found rearranged at the mRNA level in a PWS patient.10 Around 11 HERC2 copies have been identified in chromosomes 15 and 16, but most of them are on 15q11-q13.14
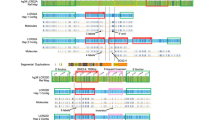
15q11-q14 chromosome rearrangements, breakpoint regions, and LCR15s clusters and copies. BP1 to BP5 breakpoints and class I (BP1) and class II (BP2) PWS/AS deletion types are shown. The (BP) located distally to marker D15S144, which has also been involved in rearrangements,22 has not been assigned a number. Contig and marker content of the 15q11-q14 region are shown. Reference markers previously reported in the definition of the breakpoint regions are shown in bold. The open rectangles indicate the HERC2-containing duplicon covered regions (not all of the markers included within these duplicons are shown). The D15S17 marker is included within the GLP genomic region. (ψ) Symbol indicates pseudogenes. Abbreviations: RP11 (RPCI-11 BAC library) and CIT (California Institute of Technology BAC library). Underlined clones are those maps at HERC2-containing duplicons. According to sequence similarities <99%, clones RP11-483E23 and RP11-291O21/228M15/578F21 located at BP3 duplicons do not overlap, representing the two BP3 duplicons (A/B). An arrow indicates the position of the clone shown in Figure 4. Asterisks indicate clones located at a different position than expected from the NT_ contigs. Filled circles indicate the presence of markers based on PCR and BLASTN analysis. The relative location of the different LCR15 groups (I to VI) and copies is shown as vertical dashed boxes. The shaded region corresponds to FISH co-hybridisation on 15q24.11 The exact number of LCR15 copies located on BP3A/B is difficult to assign due to the complexity of the zone (x?). The exact number of these repeats will not be known until the complete sequence of these zones is achieved. The only locations where we have detected the D15S17 marker (a portion of the GLP genomic sequence) are BP2, BP3A/B and between RP11-540B6 and RP5-1086D14 clones. For the rest of the LCR15 clusters the presence of other GLP sequences is indicated (x number of copies). Distances between duplicons on the NT_ contigs are not scaled. Putative duplicon orientations inferred from dot alignments are shown below.
BP2 refers to breakpoint site of class II PWS/AS patients where proximal microsatellite markers D15S18, D15S541 and D15S542 are not deleted. This is present in around half of the cases. In contrast, class I patients present a deletion of these markers and the respective breakpoint has been named BP1, being more proximal than BP2. These two breakpoint regions have been shown to be involved in similar frequencies in small inv dup(15) rearrangements and also in 15q11-q14 duplications and triplications.4,9,15,16,17,18 Three distal breakpoints have been described in 15q11-q14 rearrangements. BP3, located at 15q13, is the most common distal breakpoint, accounting for the majority of cases of PWS/AS deletions and inv dup(15). The localisation of this breakpoint was established between markers D15S12 and D15S24.19,20,21 Two other breakpoint regions named BP4 and BP5 have been mapped distally to BP3, between markers D15S24 and D15S144, and seem to be implicated in cases of large 15q11-q14 duplications or triplications.4,5,9,18,20,21 These distal regions appear to be more complex since breakpoint variability has been described (BP located distal to D15S144).9,22 These observations would suggest the existence of different repeat sequences involved in rearrangements affecting these regions.
We describe here the identification of several copies of chromosome 15 low-copy repeats (LCR15s) or duplicons at the 15q11-q14 regions of rearrangements. We reveal that the LCR15s have sequence elements in common with the HERC2-containing duplicons. The specific distribution and clustering of the LCR15s on 15q11-q14 could provide a molecular basis to understanding the rearrangements affecting these regions. This hypothesis is supported by the observation that other copies of LCR15s are located flanking a relatively common 15q24-q26 genomic mutation identified by our group.23
The results presented here are based on the analysis of the public human sequence24 and on experimental data. The non-overlapping contigs for the 15q11-q14 breakpoint and LCR15 clusters shown in Figure 1 were built through the analysis of public raw data of each genomic clone sequence (not NT_contigs) and anchored using single copy markers derived from public or published 15q11-q14 maps.10,11,25,26,27,28,29 Subsequently, the contig was compared with the data present on NT_contigs. The experimental data obtained have allowed us to manage the unfinished status of the human sequence and the putative errors derived from the draft assembly of nearly identical sequence segmental duplications or duplicons.
Materials and methods
Cell cultures and DNA samples
PWS/AS cell lines (repository nos. GM11382, GM11385, GM11404 and GM11515) were purchased from the NIGMS Genetic Mutant Cell Repository. Seven additional PWS/AS cell cultures and DNA samples were obtained from patients attending at the Corporació Parc Taulí (CPT, Sabadell, Spain). Class I or class II PWS/AS deletion types were determined through D15S18 analysis.15 Class I deletions were revealed in one GM sample (GM11515) and four samples from the CPT. Macaca fasciluaris peripheral blood lymphocytes were kindly supplied by Dr Javier Guillén. Gorilla gorilla lymphoblasts were purchased from the ECACC General Cell Collection [EB (JC) cell line].
Genomic clones and PCR analysis
The RPC1-11 BAC library was purchased by the Resource Center within the German Human Genome Project (RZPD). Filters were screened by radioactive hybridisation (Megaprime DNA labelling system, Amersham, UK) using the REP471 probe.30 RPCI-11 clones identified through the analysis of public databases but not from the initial hybridisation of this library were also purchased from the RZPD center. Positive hybridisation clones were ordered, colony isolated, BAC-DNA extracted by the alkaline lysis method and subsequently PCR analysed. REP471 amplicon sets were designed from the LCR15 sequence on 15q26.1 (RP11-697E2; GenBank accession no. AC058820): REP471 amplicon;30 F1-R1 amplicon, F1 5′-gaagcaggctatagtatccaac-3′ and R1 5′-ctacctgctgtatgccagaggttc-3′, and Rc-R1 amplicon, using forward primer Rc 5′-gacttgcaatcatgaaaacca-3′. The F1-R1 fragment (974 bp) includes the REP471 amplicon (135 bp) and the Rc-R1 amplicon (369 bp), which overlap at the Rc primer sequence (it is identical to the reverse primer sequence of the REP471 amplicon). All the clones studied that showed F1-R1 amplification also showed Rc-R1 amplification. Designed primer sets in order to obtain probes 1, 3 and 5 were: probe 1 F 5′-tgtttagttcatggtcagacg-3′ and R 5′-ttagatcacattactctctgg-3′; probe 3, F 5′-ttgctgactccagtcatg-3′ and R 5′-ttgtaaccaatacaaggactg-3′; and probe 5, F 5′-ttggaaacgccttgagaac-3′ and R 5′-cgtgttcctgtacagttcc-3′. Hybridisation conditions were standard in Church's buffer followed by stringency washes to 0.25–0.1×SSC and 0.1% SDS at 60–65°C for 30 min. Primer sets (purchased from Life Technologies) for 15q11-q14 single copy markers were those found at GDB database. SSCP analysis was performed on 12.5% polyacrylamide gels and silver staining (CleanGel DNA Analysis kit, Pharmacia). PCR agarose gel-purified fragments (QIAGEN) were sequenced according to the Big-Dye™ Terminator RR Mix protocol and analysed on an ABI 377 automated sequencer (Applied Biosystems, Inc.).
Bioinformatic resources and sequence analysis
BLASTN31 and NIX (Williams GW Woollard PM and Hingamp P: ‘NIX: A Nucleotide Identification system at the HGMP-RC’, http://www.hgmp.mrc.ac.uk/NIX/) programs were used to analyse public databases. Large genomic sequences were aligned by PipMaker and MultiPipMaker programs32 (MultiPipMaker kindly supported by Dr Webb Miller). Relatively short sequences were aligned with the CLUSTAL W program.33
FISH analysis
BAC DNA minipreparations were labelled with either biotin-16dUTP or digoxigenin-11dUTP (Boehringer Mannheim) by standard nick-translation reaction and FISH protocol was performed as described elsewhere.34 Slides were studied under a fluorescence microscope (AH3, Olympus) equipped with the appropriate filter set. Images were analysed with the Cytovision system (Applied Imaging Ltd.).
Results
LCR15 duplicon identification and localisation
Hybridisation of the RPCI-11 (RP11 abbreviated) BAC library with probe REP471, contained in duplicon LCR15 on 15q26.1,30 gave approximately 400 positive clones. From the 25×human genome coverage of this library, a first estimation of the copy number of the LCR15 duplicon was sixteen. Two hundred and eighty-one of these clones were requested and analysed by PCR for the presence of the REP471 sequence using two sets of amplicons (REP471 and F1-R1). A total of 255 clones (91%) were positive for one or two of these amplicons (+/+45%; +/−8% and −/+38%). PCR negative clones (−/−9%) could correspond to errors in reading filters from the hybridisation screening or clones containing more divergent sequences. These negative clones were not further analysed.
To further classify the positive clones we performed SSCP analysis of the REP471 and F1-R1 amplicons. We subsequently sequenced the PCR products showing different SSCP patterns allowing a classification of the clones in at least 22 groups (Table 1). Since this classification was based on PCR and SSCP analysis of a part of the LCR15 sequence, the 22 groups probably represent an underestimation of the total LCR15 copy number in the human genome. Nineteen of these 22 groups were localised on chromosome 15. Interestingly four of the groups map on 15q11-q14, one on 15q21-q22, eight on 15q24, three on 15q25 and two on 15q26. Some probes gave multiple signals by FISH and several clones gave additional signals on 15q24 probably due to the high LCR15 copy number in this region (unpublished results). The analysis of a panel of cell hybrid DNAs containing single human chromosomes35 revealed the existence of REP471 amplicons on chromosomes 2 (GenBank accession no. AJ306999), 7 (group 14) (GenBank accession no. AJ306998), 12 (GenBank accession no. AJ307000) and Y (group 7) (GenBank accession nos. AJ306997, AJ307001 and AJ307003).
LCR15 duplicon sequences were also searched by BLASTN analysis against public databases. As queries we used sequences from clones RP11-2M12 (GenBank accession no. AC013486) and RP11-697E2 (GenBank accession no. AC058820), which contain LCR15 elements that we have previously localised on 15q24 and 15q26.30 Positive sequences (cut off at >e-100, representing approximately 90 different clones) were subsequently BLASTN analysed using the NIX program through all the GenBank databases. The results of these analyses were in agreement with our previous classification and show that LCR15s are mainly distributed along chromosome 15, in five different regions: 15q11-q14, 15q21-q22, 15q24, 15q25 and 15q26. Each of these regions contains different LCR15 copies, as determined by the presence of single copy markers mapping at different locations. For example, on 15q11-q14 we have found the most proximal marker NIB1540 in the LCR15-containing clone RP11-26F2, marker D15S543, which is located distally to the BP2 duplicon, in the LCR15-containing clones RP11-228G18 and RP11-757E13, or the most distal KCC3A gene in the LCR15-containing clone RP11-122P18 (Figure 1).
The estimated total number of LCR15 related sequences in the human genome is of at least 36 copies. Most LCR15s (27 copies) belong to chromosome 15, of which at least 13 are from the 15q11-q14 region (Figure 1 and Table 1). At least nine LCR15 related sequences have been annotated to other chromosomes (2, 5, 7, 9, 10, 12, 16, 19 and Y), but the number of clones LCR15 positive clones for these other locations is smaller (up to three clones each, not shown) compared to those on chromosome 15.
Through sequence alignments we estimated the LCR15 length extent in approximately 15 kb on 15q11-14 and approximately 60 kb (without HERC2 similarities) on 15q25-q26 and chromosome Y (>95% sequence identity between them, not shown). LCR15 duplicons contain several non-processed or pseudogene sequences. High similarities to DNM1, CHRNA7, SH3P18 and GLP genes were detected in different duplicons. The length of the genomic region that could be covered by expressed sequences is of about 15 kb. The GLP BLASTN search against the EST database division revealed a large number of expressed sequences that when aligned show the existence of at least 15 different overlapping sequences. Similar results were obtained with other sequences within the LCR15s, as for example for DNM1, which is transcribed in some LCR15s in the opposite direction with respect to GLP (not shown). The GLP pseudogene is the most representative sequence of LCR15 duplicons, in agreement with the results of the screening of the RPCI-11 BAC library with probe REP471, containing GLP. The differences in the extent of LCR15 sequences are in agreement with the distinct FISH patterns detailed on Table 1 when LCR15-containing clones were used as probes.
Duplicons mapping to 15q11-q14 sites of rearrangements
The identification of LCR15-containing RPCI-11 clones, BLASTN analysis of the respective public sequences, PCR and FISH analysis allow us to identify six LCR15 clusters on 15q11-q14 (Figure 1). The assigned positions were revealed by PCR using single copy markers not included within the 15q11-q14 segmental duplications or any other duplicated sequences. The LCR15 clusters were divided in the different paralogous copies through sequence analysis that showed specific sequence differences. FISH analysis of class I and class II deletion samples allowed us to confirm the existence of LCR15 clusters located distally and proximally to the respective deletions. The LCR15 15q11-q14 constructed contig was then compared with the public NT_contigs of the region. The results confirmed the locations assigned except for proximal clones RP11-452L16 and RP11-509A17. These clones are contained within the NT–010362 contig (mapped distally, at BP2/3). However, our analysis indicates that these clones could map more proximal. They contain neurofibromatosis type 1 pseudogene sequences, which are known to be located close to the 15q centromer,25 and also present sequence similarities with other human chromosome pericentromeric regions (1q, 2p, 16p, 17p and 22q11), as has also been described for 15q pericentromer.36 The putative misassignment of these clones within the NT_010362 contig is probably due to the fact that clone RP11-509A17 contain HERC2 similarities (but with <99% sequence identity to other BP2/3 clones). The proximal position of these clones in our contig could suggest that additional HERC2-containing duplicons could map more centromeric than RP11-26F2 (Figure 1).
Multi-alignment analysis of raw data from the public sequence of genomic clones or NT_contigs shown that the previously described HERC2 duplicons10,11 and LCR15 duplicons share the genomic region containing the GLP gene (Figure 2). Therefore, as the REP471 probe is included within this region, through the present analysis we have also identified the HERC2-containing duplicons at BP2 and BP3, but also at least one duplicon copy proximal to them, probably representing BP1. Furthermore, other LCR15 that contain GLP sequences but not HERC2 sequences were identified (Figures 1 and 2). This diversification of duplicons was also suggested through hybridisation analysis. High copy numbers for HERC2 and GLP sequences, but not for other duplicon specific regions, in the human genome were observed (Figure 3A). The pip and dot-plot analysis of NT_draft sequences also allowed us to suggest the duplicon orientations on 15q11-q14 (Figure 1). Nonetheless, these orientations should be taken with caution as they are based on the unfinished status of the public human genome sequence.
Multiple sequence alignment of the NT_024668 contig sequence (377,925 kb) on BP1 (15q11) against other LCR15-containing contigs on 15q12-q14 and 15q24: overview of the NT_024668 sequence (A); detail of the duplicon-containing region (B). The locations and extensions of markers, HERC2 and GLP sequences are indicated. Since there is only one uninterrupted overlapping region, the NT_024668 contig only contains one duplicon element. The appearance of different alignments (different values of identity under the same regions) within the other contigs compared reveals the existence of more than one paralogous segment within them. Dashed lines indicate the location of the designed probes and the three different repeats revealed by the F1-R1 sequence analysis.
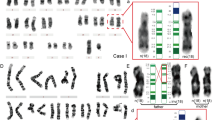
(A) Southern hybridisation results for three probes designed for the multiple alignment (Figure 2). Lane 1: GM11515; lane 2: GM11385; lane 3: control sample from the general population; and lane 4: GM11404. Arrows indicate the additional fragments in a high hybridisation background. (B) REP471 Southern hybridisation analysis of DNA from human (Hs) and non-human primates samples (Gg, Gorilla gorilla; Mm, Macaca fascicularis).
Through the BLASTN and FISH analysis of LCR15-containing 15q11-q14 clones we detected similarities with chromosome 16p11. These clones also contained HERC2 similarities. Copies of HERC2 have been described on chromosome 16 pericentromeric region.11,13,14 Sequence alignments show that the sequence responsible for this 16p11 similarity has a length of 13 kb. This HERC2-containing 16p11 duplicon is at 35 kb from another unrelated segmental duplication included within the same public clone sequence (GenBank accession no. AC002041).36
The presence of several LCR15s at the end of the BP2-HERC2 duplicon explains previous FISH reports giving signals on both 15q11-q13 and 15q24.11 Two of these copies correspond to LCR15 groups 13 and 20, contained in clone RP11-13024 (GenBank accession no. AC016033) (Table 1). Since clone 147B6 (group 19) is positive for markers D15S15, D15S17 and 363H3L (not shown), the third LCR15 copy probably corresponds to this group.
Clone RP5-1086D14 (GenBank accession no. AC004460), within cluster IV, was positive for the 3′ end of the HERC2 gene as described by Ji et al14 but not for other HERC2-containing duplicon markers. This distal region to BP3 is highly rich in partially duplicated genes (CHRNA7, MPP10, APBA2 and ACTC). Interestingly, sequence similarities with the CHRNA7 gene are also present in other LCR15 on chromosome 15q (not shown). These similarities include the previously reported duplicated sequences CHRNA7-DR1 and DR2 (GenBank accession nos. AF029838 and AF029839),29 suggesting that the LCR15 duplicon could have participated in the CHRNA7 partial duplication.
Duplicon analysis in PWS/AS 15q deletion patients
The existence of two different classes of 15q11-q13 deletions allowed us, by FISH analysis, to confirm the presence of LCR15 clusters on both sides of these rearrangements. For this purpose, we hybridised class I and II deleted chromosomes with different LCR15-containing clones: RP11-26F2 on BP1; RP11-13O24 and RP11-757E13 on BP2; RP11-291O21 and RP11-483E23 on BP3A/B; RP11-38E12 and RP11-40J8 on cluster IV; RP11-540B6 and RP5-1086D14 on cluster V; and RP11-122P18 and RP11-438P7 on cluster VI.
FISH analysis of class I samples (GM11515 and four additional samples obtained from the CPT) revealed a weaker signal on the deleted chromosome using probe RP11-26F2 (Figure 4). This observation suggests that the proximal breakpoint of this rearrangement could be located within the sequence of this clone, proximal to marker D15S18.
FISH hybridisation results of probe RP11-26F2 (BP1; Figure 1) on metaphase and interphase chromosomes from a class I deletion patient with AS (A); interphase chromosomes from the same patient hybridised with RP11-26F2 (green) and RP11-40J8 control probe (red; mapped on 15q13, see Figure 1; the metaphase chromosomes are not shown due to the proximity of the probes) (B); and class II deletion patient (C).
Analysis with probe 1 (HERC2 genomic region) revealed an additional fragment in two different DNA restriction digestions from the AS GM11404 cell line (Figure 3A). These extra fragments should correspond to a junction fragment due to the deletion affecting BP2 or BP3 regions in this patient or could represent a sequence polymorphism at one of the duplicated sequences detected by the probe. Amos-Landgraf et al10 have previously shown a possible HERC2 rearranged transcript in a PWS patient.
LCR15 analysis in non-human primates
Southern hybridisation results using the REP471 probe demonstrate the relatively recent evolutionary origin of the LCR15 sequences. This analysis was carried out with DNA from an Old World monkey (Macaca fascicularis) and one great ape species (Gorilla gorilla). A large number of LCR15 copies, albeit lower than in humans, were detected (Figure 3B). Interestingly, some of the fragments are shared by the three species compared here and in order to divergence from humans (approximately 20 Mya from M. fascicularis and approximately 7 Mya from G. gorilla).
Database search for orthologous mouse GLP sequences only identified one mouse genomic clone (RP23-170C15; GenBank accession no. AC068494). This clone contains the dynamin 1 (Dnm1) mouse gene on chromosome 2p. This region is syntenic with the human chromosome 9q33-q34, where we have also identified LCR15 similarities. Since there is only one mouse Dnm1 sequence, it could be postulated that the first human LCR15 related sequence originated from 9q33-q34, where interestingly human DNM1 and GOLGA2 (golgi autoantigen A2) genes are closely located.
Discussion
Low copy repeat sequences containing non-processed genes or pseudogenes are known as ‘duplicons’.37 Duplicons flank chromosome regions that undergo different types of human chromosomal rearrangements, such as deletions, duplications and inversions, which could occur through unequal homologous recombination events. These rearrangements seem to be promoted by the high sequence similarity between the duplicon copies and the common presence of non-processed or pseudogene expressed sequences within them.38,39
We reveal here the identification and localisation of duplicon elements in the six breakpoint regions described where 15q11-q14 rearrangements occur. While duplicons characterised by the presence of HERC2 pseudogenes have been described for two of the defined breakpoints (BP2 and BP3)10,11 duplicon sequences have not been detected for the rest of 15q11-q14 BPs. Although the breakpoint regions described at the distal LCR15s positions (clusters IV, V and VI) are >1 Mb in size and although the LCR15 extent in these regions is of approximately 15 kb, the presence of clusters (now extending the repeats to approximately 30–60 kb) at these particular locations could suggest a role in the rearrangements affecting these regions. Although a direct correlation seems to exist between the chromosome rearrangement size and the mediating duplicon extent,38,39 it is possible that other duplicon features could have an important role in promoting rearrangements. Nonetheless, the frequency of 15q11-q13 deletions, possibly involving large HERC2-containing duplicons, is higher than the described frequency for the large duplications where the LCR15 duplicons could be involved. The duplicon complexity of the 15q11-q14 region and the different rearrangement types that could be originated from them have a precedent on human chromosome 22q11 region.40,41 In addition to the putative role of LCR15 duplicons on mediating 15q11-q14 rearrangements, our group has identified other copies of LCR15s flanking a 15q24-q26 genomic mutation.23
According to what has been stated above, we suggest here the delimitation of a class I proximal breakpoint in a newly identified duplicon-containing clone. However, as this result is only based on FISH hybridisation analysis of a few class I samples, a detailed FISH and molecular analysis of a larger number of these patients is required.
Other considerations could add more complexity to the 15q11-q14 region and rearrangements. Misalignment and crossing-over between different duplicons located on the three distal LCR15 clusters could lead to not yet detected submicroscopic duplications or deletions associated to disease traits linked to these regions, as for example a neurophysiological deficit present in schizophrenia42 or autism.26 In addition, since LCR15s appear in clusters, it could be possible that a variable number of copies and/or orientations exist at specific positions in some chromosomes. These putative repeat differences could be associated to a lesser or higher susceptibility to 15q11-q14 chromosome rearrangements, in such a way as has been described for three different recurrent chromosome 8p rearrangements.43
Surprisingly, HERC2-containing duplicons and LCR15 duplicons are partially related since they share GLP gene sequences, which are characteristic of all LCR15s. Aside from chromosome 15q, we describe here that LCR15 copies also lie on chromosome Y. Detailed analysis of these copies has located them on Yq11.22, flanking the DYS7 marker (not shown). Interestingly, this marker has been reported to be recurrently deleted and duplicated.44 Recently, these golfin-related duplicons have been described associated to the AZFc region.45
The LCR15 duplicon has a large number of copies, as compared to other duplicons described so far. Interestingly most of the LCR15 copies are located on chromosome 15. Our observation of high clustering of LCR15 in discrete and large regions of the chromosome suggests that LCR15 duplicon could be considered as a ‘mobile’ element in the evolution of this chromosome. Although other types of repeated sequences such as LINE elements show high mobility rates,46 the mobility capacity of duplicons is unknown. The common presence of pseudogene sequences within LCR15 copies suggests a role for an open DNA/chromatin structure. This is a common feature for other described human duplicons and it has been suggested to facilitate recombination events.37,38,39
From the LCR15-containing clone classification described here, we have estimated the smallest sequence divergence to be as recent as 0.7 million years. On the other hand, the maximum values of estimated divergences are included within the primate evolution period (<63 Mya).47 Although these estimations are probably biased due to the specificity of the used PCR primers, they are also common for other described duplicon elements.14,37,39,48
In summary, we have shown here that the 15q11-q14 region contains a clustering of duplicons with a common signature. The present data clearly indicate the need to analyse not only the BP4 and BP5 in large 15q11-q14 rearrangements affecting these regions, but also the putative involvement of the proximal BP1 duplicon in class I PWS/AS patients. The complexity of chromosome regions containing duplicon elements may be difficult to assemble in sequenced regions of the genome. The draft sequences of the human genome have reported a large number of genomic regions that are duplicated within and between chromosomes.24,49 The anchored approach in sequencing the human genome will prove extremely valuable in resolving ambiguities (<99% sequence identity) due to the presence of duplicon sequences. Ultimately, the comparative analysis of the human genome with other duplicon-free genomes, as for example the mouse genome, should help with the correct assembly of the segmental duplications.
EMBL/GenBank accession numbers
AF375294, AF375295, AJ306964-AJ306972, AJ306981-AJ307003.
References
Khan NL, Wood NW . Prader-Willi and Angelman syndromes: update on genetic mechanisms and diagnostic complexities Curr Opin Neurol 1999 12: 149–154
Webb T, Clarke D, Hardy CA, Kilpatrick MW, Corbett J, Dahlitz M . A clinical, cytogenetic, and molecular study of 40 adults with the Prader-Willi syndrome J Med Genet 1995 32: 181–185
Bundey S, Hardy C, Vickers S, Kilpatrick MW, Corbett JA . Duplicon of the 15q11-13 region in a patient with autism, epilepsy and ataxia Dev Med Child Neurol 1994 36: 736–742
Browne CE, Dennis NR, Maher E et al. Inherited interstitial duplications of proximal 15q: genotype-phenotype correlations Am J Hum Genet 1997 61: 1342–1352
Cook EH Jr, Lindgren V, Leventhal BL et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication Am J Hum Genet 1997 60: 928–934
Schinzel AA, Brecevic L, Bernasconi F et al. Intrachromosomal triplication of 15q11-q13 J Med Genet 1994 31: 798–803
Cassidy S, Conroy J, Becker L, Schwartz S . Paternal triplication of 15q11-13 in a hypotonic, developmentally delayed child without Prader-Willi or Angelman syndrome Am J Med Genet 1996 62: 205–212
Long FL, Duckett DP, Billam LJ, Williams DK, Crolla JA . Triplication of 15q11-q13 with inv dup(15) in a female with developmental delay J Med Genet 1998 35: 425–428
Ungaro P, Christian SL, Fantes JA et al. Molecular characterisation of four cases of intrachromosomal triplication of chromosome 15q11-q14 J Med Genet 2001 38: 26–34
Amos-Landgraf JM, Ji Y, Gottlieb W et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints Am J Hum Genet 1999 65: 370–386
Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH . Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet 1999 8: 1025–1037
Donlon TA, Lalande M, Wyman A, Bruns G, Latt SA . Isolation of molecular probes associated with the chromosome 15 instability in the Prader-Willi syndrome Proc Natl Acad Sci USA 1986 83: 4408–4412
Ji Y, Walkowicz MJ, Buiting K et al. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities Hum Mol Genet 1999 8: 533–542
Ji Y, Rebert NA, Joslin JM, Higgins MJ, Schultz RA, Nicholls RD . Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human Genome Res 2000 10: 319–329
Knoll JH, Nicholls RD, Magenis RE et al. Angelman syndrome: three molecular classes identified with chromosome 15q11q13-specific DNA markers Am J Hum Genet 1990 47: 149–155
Christian SL, Robinson WP, Huang B et al. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients Am J Hum Genet 1995 57: 40–48
Huang B, Crolla JA, Christian SL et al. Refined molecular characterization of the breakpoints in small inv dup(15) chromosomes Hum Genet 1997 99: 11–17
Robinson WP, Horsthemke B, Leonard S et al. Report of the Third International Workshop on Human Chromosome 15 Mapping 1996. October 25–27, 1996 in Vancouver B.C., Canada Cytogenet Cell Genet 1997 76: 1–13
Cheng SD, Spinner NB, Zackai EH, Knoll JH . Cytogenetic and molecular characterization of inverted duplicated chromosomes 15 from 11 patients Am J Hum Genet 1994 55: 753–759
Crolla JA, Harvey JF, Sitch FL, Dennis NR . Supernumerary marker 15 chromosomes: a clinical, molecular and FISH approach to diagnosis and prognosis Hum Genet 1995 95: 161–170
Wandstrat AE, Leana-Cox J, Jenkins L, Schwartz S . Molecular cytogenetic evidence for a common breakpoint in the largest inverted duplications of chromosome 15 Am J Hum Genet 1998 62: 925–936
Robinson WP, Dutly F, Nicholls RD et al. The mechanisms involved in formation of deletions and duplications of 15q11-q13 J Med Genet 1998 35: 130–136
Gratacòs M, Nadal M, Martín-Santos R et al. A polymorphic genomic duplication on human chromosome 15 is a susceptibility factor for panic and phobic disorders Cell 2001 106: 367–379
Lander ES, Linton LM, Birren B et al. Initial sequencing and analysis of the human genome Nature 2001 409: 860–921
Ritchie RJ, Mattei MG, Lalande M . A large polymorphic repeat in the pericentromeric region of human chromosome 15q11-q13 contains three partial gene duplications Hum Mol Genet 1998 7: 1253–1260
Maddox LO, Menold MM, Bass MP et al. Autistic disorder and chromosome 15q11-q13: construction and analysis of a BAC/PAC contig Genomics 1999 62: 325–331
Christian SL, Bhatt NK, Martin SA et al. Integrated YAC contig map of the Prader-Willi/Angelman region on chromosome 15q11-q13 with average STS spacing of 35 kb Genome Res 1998 8: 146–157
Morton CC, Christian SL, Donlon TA et al. Report of the fourth international workshop on human chromosome 15 mapping 1997 Cytogenet Cell Genet 1999 84: 12–21
Gault J, Robinson M, Berger R et al. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics 1998 52: 173–185
Pujana MA, Nadal M, Gratacòs M et al. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26 Genome Res 2001 11: 98–111
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . Basic local alignment search tool J Mol Biol 1990 215: 403–410
Schwartz S, Zhang Z, Frazer KA et al. PipMaker – a web server for aligning two genomic DNA sequences Genome Res 2000 10: 577–586
Thompson JD, Higgins DG, Gibson TJ . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res 1994 22: 4673–4680
Nadal M, Moreno S, Pritchard M, Preciado MA, Estivill X, Ramos-Arroyo MA . Down syndrome: characterisation of a case with partial trisomy of chromosome 21 owing to a paternal balanced translocation (15;21) (q26;q22.1) by FISH J Med Genet 1997 34: 50–54
Dubois BL, Naylor SL . Characterization of NIGMS human/rodent somatic cell hybrid mapping panel 2 by PCR Genomics 1993 16: 315–319
Horvath JE, Schwartz S, Eichler EE . The mosaic structure of human pericentromeric DNA: a strategy for characterizing complex regions of the human genome Genome Res 2000 10: 839–852
Eichler EE . Masquerading repeats: paralogous pitfalls of the human genome Genome Res 1998 8: 758–762
Lupski JR . Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits Trends Genet 1998 14: 417–422
Ji Y, Eichler EE, Schwartz S, Nicholls RD . Structure of chromosomal duplicons and their role in mediating human genomic disorders Genome Res 2000 10: 597–610
Shaikh TH, Kurahashi H, Saitta SC et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis Hum Mol Genet 2000 9: 489–501
Edelmann L, Pandita RK, Spiteri E et al. A common molecular basis for rearrangement disorders on chromosome 22q11 Hum Mol Genet 1999 8: 1157–1167
Freedman R, Coon H, Myles-Worsley M et al. Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus Proc Natl Acad Sci USA 1997 94: 587–592
Giglio S, Broman KW, Matsumoto N et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements Am J Hum Genet 2001 68: 874–883
Jobling MA, Samara V, Pandya A et al. Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males Hum Mol Genet 1996 5: 1767–1775
Kuroda-Kawaguchi T, Skaletsky H, Brown LG et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men Nature Genet 2001 29: 279–286
Pickeral OK, Makalowski W, Boguski MS, Boeke JD . Frequent human genomic DNA transduction driven by LINE-1 retrotransposition Genome Res 2001 10: 411–415
Goodman M . The genomic record of Humankind's evolutionary roots Am J Hum Genet 1999 64: 31–39
Keller MP, Seifried BA, Chance PF . Molecular evolution of the CMT1A-REP region: a human- and chimpanzee-specific repeat Mol Biol Evol 1999 16: 1019–1026
Venter JC, Adams MD, Myers EW et al. The sequence of the human genome Science 2001 291: 1304–1351
Acknowledgements
We thank Dr Javier Guillén for providing peripheral blood lymphocytes from Macaca fascicularis. We thank Helena Kruyer for assistance in preparing the manuscript. This work was supported by La Marató de TV3 (98/1810), the European Union (BMH4-CT97-2284) and the Spanish Government (CICYT; SAF99-0092-CO2-01). MA Pujana is a fellow of the Centre de Regulació Genòmica. L Armengol is a fellow of the Comissió Interdepartamental de Recerca i Innovació Tecnològica of the Generalitat de Catalunya.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pujana, M., Nadal, M., Guitart, M. et al. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet 10, 26–35 (2002). https://doi.org/10.1038/sj.ejhg.5200760
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200760
Keywords
This article is cited by
-
Genetic subtypes and phenotypic characteristics of 110 patients with Prader-Willi syndrome
Italian Journal of Pediatrics (2022)
-
A rare familial rearrangement of chromosomes 9 and 15 associated with intellectual disability: a clinical and molecular study
Molecular Cytogenetics (2021)
-
First report of two successive deletions on chromosome 15q13 cytogenetic bands in a boy and girl: additional data to 15q13.3 syndrome with a report of high IQ patient
Molecular Cytogenetics (2019)
-
CYFIP1 overexpression increases fear response in mice but does not affect social or repetitive behavioral phenotypes
Molecular Autism (2019)
-
Phenotypic association of 15q11.2 CNVs of the region of breakpoints 1–2 (BP1–BP2) in a large cohort of samples referred for genetic diagnosis
Journal of Human Genetics (2019)