Abstract
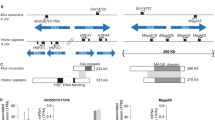
Mammalian sex determination is governed by the presence of the sex determining region Y gene (SRY) on the Y chromosome1. Familial cases of SRY-negative XX sex reversal are rare in humans, often hampering the discovery of new sex-determining genes2,3. The mouse model is also insufficient to correctly apprehend the sex-determination cascade, as the human pathway is much more sensitive to gene dosage4,5,6. Other species might therefore be considered in this respect7. In goats, the polled intersex syndrome (PIS) mutation associates polledness and intersexuality8,9. The sex reversal affects exclusively the XX individuals in a recessive manner, whereas the absence of horns is dominant in both sexes. The syndrome is caused by an autosomal gene located at chromosome band 1q43 (ref. 9), shown to be homologous to human chromosome band 3q23 (ref. 10). Through a positional cloning approach, we demonstrate that the mutation underlying PIS is the deletion of a critical 11.7-kb DNA element containing mainly repetitive sequences. This deletion affects the transcription of at least two genes: PISRT1, encoding a 1.5-kb mRNA devoid of open reading frame (ORF), and FOXL2, recently shown to be responsible for blepharophimosis ptosis epicanthus inversus syndrome (BPES) in humans11. These two genes are located 20 and 200 kb telomeric from the deletion, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Koopman, P., Gubbay, J., Vivian, N., Goodfellow, P. & Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121 (1991).
Krob, G., Braun, A. & Kuhnle, U. True hermaphroditism: geographical distribution, clinical findings, chromosomes and gonadal histology. Eur. J. Pediatr. 153, 2–10 (1994).
McElreavey, K., Vilain, E., Abbas, N., Herskowitz, I. & Fellous, M. A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc. Natl Acad. Sci. USA 90, 3368–3372 (1993).
Swain, A., Narvaez, V., Burgoyne, P., Camerino, G. & Lovell-Badge, R. Dax1 antagonizes Sry action in mammalian sex determination. Nature 391, 761–767 (1998).
Luo, X., Ikeda, Y. & Parker, K.L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77, 481–490 (1994).
Kreidberg, J.A. et al. WT-1 is required for early kidney development. Cell 74, 679–691 (1993).
Vaiman, D. & Pailhoux, E. Mammalian sex reversal and intersexuality: deciphering the sex-determination cascade. Trends Genet. 16, 488–494 (2000).
Asdell, S.A. The genetic sex on intersexual goats and a probable linkage with the gene for hornlessness. Science 99, 124 (1944).
Vaiman, D. et al. Genetic mapping of the autosomal region involved in XX sex-reversal and horn development in goats. Mamm. Genome 7, 133–137 (1996).
Vaiman, D. et al. High-resolution human/goat comparative map of the goat polled/intersex syndrome (PIS): the human homologue is contained in a human YAC from HSA3q23. Genomics 56, 31–39 (1999).
Crisponi, L. et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nature Genet. 27, 159–166 (2001).
Schibler, L., Cribiu, E.P., Oustry-Vaiman, A., Furet, J.P. & Vaiman, D. Fine mapping suggests that the goat Polled Intersex Syndrome and the human Blepharophimosis Ptosis Epicanthus Syndrome map to a 100-kb homologous region. Genome Res. 10, 311–318 (2000).
Smale, S.T. & Baltimore, D. The “initiator” as a transcription control element. Cell 57, 103–113 (1989).
Lawson, C.T. et al. Definition of the blepharophimosis, ptosis, epicanthus inversus syndrome critical region at chromosome 3q23 based on the analysis of chromosomal anomalies. Hum. Mol. Genet. 4, 963–967 (1995).
De Baere, E. et al. Identification of BPESC1, a novel gene disrupted by a balanced chromosomal translocation, t(3;4)(q23;p15.2), in a patient with BPES. Genomics 68, 296–304 (2000).
Praphanphoj, V. et al. Molecular cytogenetic evaluation in a patient with a translocation (3;21) associated with blepharophimosis, ptosis, epicanthus inversus syndrome (BPES). Genomics 65, 67–69 (2000).
Koopman, P. Sry and Sox9: mammalian testis-determining genes. Cell Mol. Life Sci. 55, 839–856 (1999).
Arango, N.A., Lovell-Badge, R. & Behringer, R.R. Targeted mutagenesis of the endogenous mouse Mis gene promoter: in vivo definition of genetic pathways of vertebrate sexual development. Cell 99, 409–419 (1999).
Giese, K., Cox, J. & Grosschedl, R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69, 185–195 (1992).
Giese, K., Pagel, J. & Grosschedl, R. Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc. Natl Acad. Sci. USA 91, 3368–3372 (1994).
Aasen, E. & Medrano, J.F. Amplification of the ZFY and ZFX genes for sex identification in humans, cattle, sheep and goats. Biotechnology 8, 1279–1281 (1990).
Vaiman, D. et al. A genetic linkage map of the male goat genome. Genetics 144, 279–305 (1996).
Acknowledgements
The authors wish to acknowledge the work of R. Faugeras for sequencing and the excellent technical assistance of B. Jego. We thank J. Hervieu and J.F. Alkombre for help with goat breeding and management. We wish to acknowledge J.L. Vilotte for critical reading of the manuscript. This work was carried out with INRA AIP grants “Sex autosomal determination in goats and pigs” and “Genome and function”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pailhoux, E., Vigier, B., Chaffaux, S. et al. A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat Genet 29, 453–458 (2001). https://doi.org/10.1038/ng769
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng769
This article is cited by
-
Genome-wide detection of copy number variation in American mink using whole-genome sequencing
BMC Genomics (2022)
-
Genome-wide association study reveals 14 new SNPs and confirms two structural variants highly associated with the horned/polled phenotype in goats
BMC Genomics (2021)
-
A genome-wide analysis of copy number variation in Murciano-Granadina goats
Genetics Selection Evolution (2020)
-
Whole genome resequencing of the Iranian native dogs and wolves to unravel variome during dog domestication
BMC Genomics (2020)
-
Genome-wide analysis of Chongqing native intersexual goats using next-generation sequencing
3 Biotech (2019)