Abstract
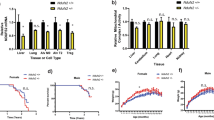
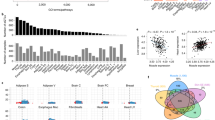
Common mitochondrial DNA (mtDNA) haplotypes in humans and mice have been associated with various phenotypes, including learning performance and disease penetrance. Notably, no influence of mtDNA haplotype in cell respiration has been demonstrated. Here, using cell lines carrying four different common mouse mtDNA haplotypes in an identical nuclear background, we show that the similar level of respiration among the cell lines is only apparent and is a consequence of compensatory mechanisms triggered by different production of reactive oxygen species. We observe that the respiration capacity per molecule of mtDNA in cells with the NIH3T3 or NZB mtDNA is lower than in those with the C57BL/6J, CBA/J or BALB/cJ mtDNA. In addition, we have determined the genetic element underlying these differences. Our data provide insight into the molecular basis of the complex phenotypes associated with common mtDNA variants and anticipate a relevant contribution of mtDNA single nucleotide polymorphisms to phenotypic variability in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ruiz-Pesini, E. et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Hum. Genet. 67, 682–696 (2000).
Montiel-Sosa, F. et al. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene 368C, 21–27 (2006).
Tanaka, M., Gong, J.S., Zhang, J., Yoneda, M. & Yagi, K. Mitochondrial genotype associated with longevity. Lancet 351, 185–186 (1998).
De Benedictis, G. et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 13, 1532–1536 (1999).
Niemi, A.K. et al. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 112, 29–33 (2003).
Brown, M.D. et al. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup J. Hum. Genet. 110, 130–138 (2002).
Howell, N., Herrnstadt, C., Shults, C. & Mackey, D.A. Low penetrance of the 14484 LHON mutation when it arises in a non-haplogroup J mtDNA background. Am. J. Med. Genet. A. 119, 147–151 (2003).
Wallace, D.C., Ruiz-Pesini, E. & Mishmar, D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harb. Symp. Quant. Biol. 68, 479–486 (2003).
Dimauro, S. Mitochondrial medicine. Biochim. Biophys. Acta 1659, 107–114 (2004).
Ruiz-Pesini, E., Mishmar, D., Brandon, M., Procaccio, V. & Wallace, D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303, 223–226 (2004).
Johnson, K.R., Zheng, Q.Y., Bykhovskaya, Y., Spirina, O. & Fischel-Ghodsian, N. A nuclear–mitochondrial DNA interaction affecting hearing impairment in mice. Nat. Genet. 27, 191–194 (2001).
Noben-Trauth, K., Zheng, Q.Y. & Johnson, K.R. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 35, 21–23 (2003).
Jenuth, J.P., Peterson, A.C. & Shoubridge, E.A. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16, 93–95 (1997).
Roubertoux, P.L. et al. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat. Genet. 35, 65–69 (2003).
Carelli, V. et al. Respiratory function in cybrid cell lines carrying European mtDNA haplogroups: implications for Leber's hereditary optic neuropathy. Biochim. Biophys. Acta 1588, 7–14 (2002).
Battersby, B.J. & Shoubridge, E.A. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 10, 2469–2479 (2001).
Herrnstadt, C. & Howell, N. An evolutionary perspective on pathogenic mtDNA mutations: haplogroup associations of clinical disorders. Mitochondrion 4, 797–798 (2004).
Acin-Perez, R. et al. An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet. 12, 329–339 (2003).
Bai, Y. & Attardi, G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 17, 4848–4858 (1998).
Acin-Perez, R. et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 (2004).
Loveland, B., Wang, C.R., Yonekawa, H., Hermel, E. & Lindahl, K.F. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell 60, 971–980 (1990).
Bayona-Bafaluy, M.P. et al. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 31, 5349–5355 (2003).
Beckman, K.B. & Ames, B.N. The free radical theory of aging matures. Physiol. Rev. 78, 547–581 (1998).
Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95 (2002).
Lee, H.C. & Wei, Y.H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int. J. Biochem. Cell Biol. 37, 822–834 (2005).
Geromel, V. et al. Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum. Mol. Genet. 10, 1221–1228 (2001).
Lee, H.C., Yin, P.H., Chi, C.W. & Wei, Y.H. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J. Biomed. Sci. 9, 517–526 (2002).
Liu, C.S. et al. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic. Res. 37, 1307–1317 (2003).
Nulton-Persson, A.C. & Szweda, L.I. Modulation of mitochondrial function by hydrogen peroxide. J. Biol. Chem. 276, 23357–23361 (2001).
Tretter, L. & Adam-Vizi, V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of α-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 20, 8972–8979 (2000).
Gardner, P.R., Nguyen, D.D. & White, C.W. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc. Natl. Acad. Sci. USA 91, 12248–12252 (1994).
Gardner, P.R., Raineri, I., Epstein, L.B. & White, C.W. Superoxide radical and iron modulate aconitase activity in mammalian cells. J. Biol. Chem. 270, 13399–13405 (1995).
Zhao, H. et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 74, 139–152 (2004).
Prezant, T.R. et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4, 289–294 (1993).
Reid, F.M., Vernham, G.A. & Jacobs, H.T. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum. Mutat. 3, 243–247 (1994).
Guan, M.X. et al. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASerUCN precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol. Cell. Biol. 18, 5868–5879 (1998).
Finkel, T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 15, 247–254 (2003).
Chomyn, A. et al. Platelet-mediated transformation of mtDNA-less human cells: analysis of phenotypic variability among clones from normal individuals—and complementation behavior of the tRNALys mutation causing myoclonic epilepsy and ragged red fibers. Am. J. Hum. Genet. 54, 966–974 (1994).
King, M.P. & Attardi, G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503 (1989).
Gallardo, M.E. et al. m627G>A, a recurrent mutation in the human mitochondrial DNA that reduce cytochrome c oxidase activity and is associated with tumors. Hum. Mutat. 27, 575–582 (2006).
Staden, R., Beal, K.F. & Bonfield, J.K. The Staden package, 1998. Methods Mol. Biol. 132, 115–130 (2000).
Hofhaus, G., Shakeley, R.M. & Attardi, G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 264, 476–483 (1996).
Fernandez-Vizarra, E., Lopez-Perez, M.J. & Enriquez, J.A. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods 26, 292–297 (2002).
Wang, H. & Joseph, J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 27, 612–616 (1999).
Aebi, H. Catalase in vitro. Methods Enzymol. 105, 121–126 (1984).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Enriquez, J.A., Fernandez-Silva, P., Perez-Martos, A., Lopez-Perez, M.J. & Montoya, J. The synthesis of mRNA in isolated mitochondria can be maintained for several hours and is inhibited by high levels of ATP. Eur. J. Biochem. 237, 601–610 (1996).
Enriquez, J.A., Chomyn, A. & Attardi, G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat. Genet. 10, 47–55 (1995).
Acknowledgements
We thank C. Moraes and E. Shoubridge for the NZB hepatocytes; E. Ruiz-Pesini, A. Barrientos, C. López-Otin and M. Palacín for comments on the manuscript; and S. Morales for technical assistance. Our work was supported by the Spanish Ministry of Education (SAF2003-00103), the Instituto de Salud Carlos III (REDEMETH-G03/054, REDCIEN C03/06-Grupo RC-N34-3 yr ECEMECRE G03/011), the European Union (EUMITOCOMBAT-LSHM-CT-2004-503116), Group of Excellence grant DGA (B55) and Fundación Ramón Areces. R.M.-L. and R.A.-P. are supported by a predoctoral fellowship from the Spanish Ministry of Education and M.E.G. is supported by an I3P postdoctoral contract from the CSIC, Spain.
Author information
Authors and Affiliations
Contributions
This study was designed and the paper was written by P.F.-S., A.P.-M., S.R.d.C. and J.A.E.; the wild-type cybrid cell line was constructed and phenotype assessed by R.M.-L. and R.A.-P.; the TmND6ko mtDNA mutant cell line was generated and characterized by N.M.; and the mtDNA resequencing strategy was designed by, and full mtDNA sequencing performed by, M.E.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
RFLP analysis to detect the possible presence of L929 mtDNA in the transmitochondrial cell lines. (PDF 112 kb)
Supplementary Fig. 2
Protein levels for specific subunits of the mtETC complexes in the different mtDNA haplotypes. (PDF 104 kb)
Supplementary Fig. 3
ND3/ND4 relative transcript levels in the transmitochondrial cell lines. (PDF 98 kb)
Supplementary Table 1
mtDNA nucleotide differences between C57BL/6J and NZB/B1NJ. (PDF 18 kb)
Supplementary Table 2
Primer sequences. (PDF 80 kb)
Rights and permissions
About this article
Cite this article
Moreno-Loshuertos, R., Acín-Pérez, R., Fernández-Silva, P. et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet 38, 1261–1268 (2006). https://doi.org/10.1038/ng1897
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1897
This article is cited by
-
Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2
Nature Metabolism (2021)
-
Presence of male mitochondria in somatic tissues and their functional importance at the whole animal level in the marine bivalve Arctica islandica
Communications Biology (2021)
-
Common mtDNA variations at C5178a and A249d/T6392C/G10310A decrease the risk of severe COVID-19 in a Han Chinese population from Central China
Military Medical Research (2021)
-
ROS and diseases: role in metabolism and energy supply
Molecular and Cellular Biochemistry (2020)
-
Novel compound mutations in the mitochondrial translation elongation factor (TSFM) gene cause severe cardiomyopathy with myocardial fibro-adipose replacement
Scientific Reports (2019)