Abstract
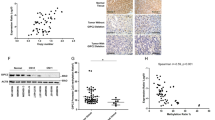
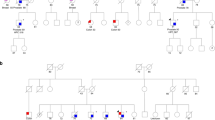
Hereditary pheochromocytoma (PCC) is often caused by germline mutations in one of nine susceptibility genes described to date1,2,3,4, but there are familial cases without mutations in these known genes. We sequenced the exomes of three unrelated individuals with hereditary PCC (cases) and identified mutations in MAX, the MYC associated factor X gene. Absence of MAX protein in the tumors and loss of heterozygosity caused by uniparental disomy supported the involvement of MAX alterations in the disease. A follow-up study of a selected series of 59 cases with PCC identified five additional MAX mutations and suggested an association with malignant outcome and preferential paternal transmission of MAX mutations. The involvement of the MYC-MAX-MXD1 network in the development and progression of neural crest cell tumors is further supported by the lack of functional MAX in rat PCC (PC12) cells5 and by the amplification of MYCN in neuroblastoma6 and suggests that loss of MAX function is correlated with metastatic potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Neumann, H.P. et al. Germ-line mutations in nonsyndromic pheochromocytoma. N. Engl. J. Med. 346, 1459–1466 (2002).
Hao, H.X. et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325, 1139–1142 (2009).
Yao, L. et al. Spectrum and prevalence of FP/TMEM127 gene mutations in pheochromocytomas and paragangliomas. J. Am. Med. Assoc. 304, 2611–2619 (2010).
Burnichon, N. et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum. Mol. Genet. 19, 3011–3020 (2010).
Hopewell, R. & Ziff, E.B. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol. Cell. Biol. 15, 3470–3478 (1995).
Kohl, N.E. et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 35, 359–367 (1983).
Mannelli, M. et al. Clinically guided genetic screening in a large cohort of Italian patients with pheochromocytomas and/or functional or nonfunctional paragangliomas. J. Clin. Endocrinol. Metab. 94, 1541–1547 (2009).
Cascón, A. et al. Genetics of pheochromocytoma and paraganglioma in Spanish patients. J. Clin. Endocrinol. Metab. 94, 1701–1705 (2009).
Favier, J. et al. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS ONE 4, e7094 (2009).
Dahia, P.L. et al. A HIF1α regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 1, 72–80 (2005).
López-Jiménez, E. et al. Research resource: transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol. Endocrinol. 24, 2382–2391 (2010).
Atchley, W.R. & Fitch, W.M. Myc and Max: molecular evolution of a family of proto-oncogene products and their dimerization partner. Proc. Natl. Acad. Sci. USA 92, 10217–10221 (1995).
Grandori, C., Cowley, S.M., James, L.P. & Eisenman, R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699 (2000).
Bousset, K., Henriksson, M., Luscher-Firzlaff, J.M., Litchfield, D.W. & Luscher, B. Identification of casein kinase II phosphorylation sites in Max: effects on DNA-binding kinetics of Max homo- and Myc/Max heterodimers. Oncogene 8, 3211–3220 (1993).
Prendergast, G.C., Hopewell, R., Gorham, B.J. & Ziff, E.B. Biphasic effect of Max on Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions. Genes Dev. 6, 2429–2439 (1992).
Ribon, V., Leff, T. & Saltiel, A.R. c-Myc does not require max for transcriptional activity in PC-12 cells. Mol. Cell. Neurosci. 5, 277–282 (1994).
Segouffin-Cariou, C. & Billaud, M. Transforming ability of MEN2A-RET requires activation of the phosphatidylinositol 3-kinase/AKT signaling pathway. J. Biol. Chem. 275, 3568–3576 (2000).
Johannessen, C.M. et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. USA 102, 8573–8578 (2005).
Qin, Y. et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat. Genet. 42, 229–233 (2010).
Zhu, J., Blenis, J. & Yuan, J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA 105, 6584–6589 (2008).
Jimenez, R.H. et al. Regulation of gene expression in hepatic cells by the mammalian Target of Rapamycin (mTOR). PLoS ONE 5, e9084 (2010).
Vaqué, J.P. et al. c-Myc inhibits Ras-mediated differentiation of pheochromocytoma cells by blocking c-Jun up-regulation. Mol. Cancer Res. 6, 325–339 (2008).
Nair, S.K. & Burley, S.K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003).
Dang, C.V., McGuire, M., Buckmire, M. & Lee, W.M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature 337, 664–666 (1989).
Teh, M.T. et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 65, 8597–8603 (2005).
Murthy, S.K., DiFrancesco, L.M., Ogilvie, R.T. & Demetrick, D.J. Loss of heterozygosity associated with uniparental disomy in breast carcinoma. Mod. Pathol. 15, 1241–1250 (2002).
Tiu, R.V. et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J. Clin. Oncol. 27, 5219–5226 (2009).
Kurosawa, K. et al. Paternal UPD14 is responsible for a distinctive malformation complex. Am. J. Med. Genet. 110, 268–272 (2002).
Wilson, M. et al. The clinical phenotype of mosaicism for genome-wide paternal uniparental disomy: two new reports. Am. J. Med. Genet. A. 146A, 137–148 (2008).
Baysal, B.E. et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 (2000).
Sambrook, J., Maniatis, T. & Fritsch, E.F. Molecular Cloning: A Laboratory Manual, 3 v. (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1989).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Simon-Sanchez, J. et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum. Mol. Genet. 16, 1–14 (2007).
Cascón, A. et al. Gross SDHB deletions in patients with paraganglioma detected by multiplex PCR: a possible hot spot? Genes Chromosom. Cancer 45, 213–219 (2006).
Astuti, D. et al. Epigenetic alteration at the DLK1–GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour. Br. J. Cancer 92, 1574–1580 (2005).
Landa, I. et al. Allelic variant at -79 (C>T) in CDKN1B (p27Kip1) confers an increased risk of thyroid cancer and alters mRNA levels. Endocr. Relat. Cancer 17, 317–328 (2010).
Acknowledgements
This work was supported in part by the Fondo de Investigaciones Sanitarias (projects PS09/00942 and P1080883 to A.C. and M.R., respectively), Mutua Madrileña (project AP2775/2008 to M.R.), FP7-Grant (ENS@T-CANCER; HEALTH-F2-2010-259735) and Innovation project INTRA-706-2 ISCIII CIBER-ER (Center for Biomedical Research on Rare Diseases). I.C.-M. holds a shuttle CIBER-ER fellowship.
Author information
Authors and Affiliations
Contributions
A.C., M.R., F.S. and G.O. conceived the project. G. Pica, P.L., R.H.-L., J.A.D., M.G.-M. and M.M. collected tumor samples. F.J.G.-A., I.C.-M. and A.C. performed next-generation sequencing analysis and filtering. I.C.-M., F.J.G.-A., F.S., I.L., L.J.L.-G., R.L., R.R.-M., D.C., A.G.-G., A.A.d.C., L.I.-P., E.T., S.B., A.M., A.G.-N. and G.R. performed additional experiments. A.C., I.C.-M., E.H. and G. Pita performed additional data analysis. A.C., I.C.-M., F.J.G.-A., M.R., C.R.-A. and J.B. wrote the manuscript. All authors approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–3. (PDF 315 kb)
Rights and permissions
About this article
Cite this article
Comino-Méndez, I., Gracia-Aznárez, F., Schiavi, F. et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet 43, 663–667 (2011). https://doi.org/10.1038/ng.861
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.861
This article is cited by
-
Wilms tumour resulting from paternal transmission of a TRIM28 pathogenic variant—A first report
European Journal of Human Genetics (2024)
-
Organ and cell-specific biomarkers of Long-COVID identified with targeted proteomics and machine learning
Molecular Medicine (2023)
-
Single-nuclei and bulk-tissue gene-expression analysis of pheochromocytoma and paraganglioma links disease subtypes with tumor microenvironment
Nature Communications (2022)
-
Overview of the 2022 WHO Classification of Familial Endocrine Tumor Syndromes
Endocrine Pathology (2022)
-
MYCN and MAX alterations in Wilms tumor and identification of novel N-MYC interaction partners as biomarker candidates
Cancer Cell International (2021)