Abstract
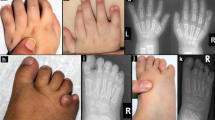
Brain malformations are individually rare but collectively common causes of developmental disabilities1,2,3. Many forms of malformation occur sporadically and are associated with reduced reproductive fitness, pointing to a causative role for de novo mutations4,5. Here, we report a study of Baraitser-Winter syndrome, a well-defined disorder characterized by distinct craniofacial features, ocular colobomata and neuronal migration defect6,7. Using whole-exome sequencing of three proband-parent trios, we identified de novo missense changes in the cytoplasmic actin–encoding genes ACTB and ACTG1 in one and two probands, respectively. Sequencing of both genes in 15 additional affected individuals identified disease-causing mutations in all probands, including two recurrent de novo alterations (ACTB, encoding p.Arg196His, and ACTG1, encoding p.Ser155Phe). Our results confirm that trio-based exome sequencing is a powerful approach to discover genes causing sporadic developmental disorders, emphasize the overlapping roles of cytoplasmic actin proteins in development and suggest that Baraitser-Winter syndrome is the predominant phenotype associated with mutation of these two genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
04 March 2012
In the version of this article initially published online, the proband from trio 1 was incorrectly identified as LP98-083 in the second paragraph of the main text. The correct identification number for this subject is LP92-083. The error has been corrected for the print, PDF and HTML versions of this article.
References
Glinianaia, S.V., Rankin, J. & Colver, A. Cerebral palsy rates by birth weight, gestation and severity in North of England, 1991–2000 singleton births. Arch. Dis. Child. 96, 180–185 (2011).
Rankin, J. et al. Congenital anomalies in children with cerebral palsy: a population-based record linkage study. Dev. Med. Child Neurol. 52, 345–351 (2010).
von Wendt, L. & Rantakallio, P. Congenital malformations of the central nervous system in a 1-year birth cohort followed to the age of 14 years. Childs Nerv. Syst. 2, 80–82 (1986).
Vissers, L.E. et al. A de novo paradigm for mental retardation. Nat. Genet. 42, 1109–1112 (2010).
Cooper, G.M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Baraitser, M. & Winter, R.M. Iris coloboma, ptosis, hypertelorism, and mental retardation: a new syndrome. J. Med. Genet. 25, 41–43 (1988).
Ramer, J.C. et al. Previously apparently undescribed syndrome: shallow orbits, ptosis, coloboma, trigonocephaly, gyral malformations, and mental and growth retardation. Am. J. Med. Genet. 57, 403–409 (1995).
Hoischen, A. et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 42, 483–485 (2010).
Hoischen, A. et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 43, 729–731 (2011).
Ng, S.B. et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 (2010).
Conrad, D.F. et al. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 43, 712–714 (2011).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107, 961–968 (2010).
O'Roak, B.J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Girard, S.L. et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 43, 860–863 (2011).
Xu, B. et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 43, 864–868 (2011).
Fryns, J.P. & Aftimos, S. New MR/MCA syndrome with distinct facial appearance and general habitus, broad and webbed neck, hypoplastic inverted nipples, epilepsy, and pachygyria of the frontal lobes. J. Med. Genet. 37, 460–462 (2000).
Rossi, M., Guerrini, R., Dobyns, W.B., Andria, G. & Winter, R.M. Characterization of brain malformations in the Baraitser-Winter syndrome and review of the literature. Neuropediatrics 34, 287–292 (2003).
Verloes, A. Iris coloboma, ptosis, hypertelorism, and mental retardation: Baraitser-Winter syndrome or Noonan syndrome? J. Med. Genet. 30, 425–426 (1993).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Scarano, E., Iaccarino, M., Grippo, P. & Parisi, E. The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos. Proc. Natl. Acad. Sci. USA 57, 1394–1400 (1967).
Buetow, K.H., Edmonson, M.N. & Cassidy, A.B. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21, 323–325 (1999).
Gupton, S.L. & Gertler, F.B. Filopodia: the fingers that do the walking. Sci. STKE 2007, re5 (2007).
Harborth, J., Elbashir, S.M., Bechert, K., Tuschl, T. & Weber, K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557–4565 (2001).
Lambrechts, A., Van Troys, M. & Ampe, C. The actin cytoskeleton in normal and pathological cell motility. Int. J. Biochem. Cell Biol. 36, 1890–1909 (2004).
Shawlot, W., Deng, J.M., Fohn, L.E. & Behringer, R.R. Restricted β-galactosidase expression of a hygromycin-lacZ gene targeted to the β-actin locus and embryonic lethality of β-actin mutant mice. Transgenic Res. 7, 95–103 (1998).
Vandekerckhove, J. & Weber, K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 126, 783–802 (1978).
Garrels, J.I. & Gibson, W. Identification and characterization of multiple forms of actin. Cell 9, 793–805 (1976).
Yao, J., Sasaki, Y., Wen, Z., Bassell, G.J. & Zheng, J.Q. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 9, 1265–1273 (2006).
Dugina, V., Zwaenepoel, I., Gabbiani, G., Clement, S. & Chaponnier, C. β and γ-cytoplasmic actins display distinct distribution and functional diversity. J. Cell Sci. 122, 2980–2988 (2009).
Höfer, D., Ness, W. & Drenckhahn, D. Sorting of actin isoforms in chicken auditory hair cells. J. Cell Sci. 110, 765–770 (1997).
Belyantseva, I.A. et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA 106, 9703–9708 (2009).
Bunnell, T.M. & Ervasti, J.M. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton (Hoboken) 67, 564–572 (2010).
Nunoi, H. et al. A heterozygous mutation of β-actin associated with neutrophil dysfunction and recurrent infection. Proc. Natl. Acad. Sci. USA 96, 8693–8698 (1999).
Procaccio, V. et al. A mutation of β-actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am. J. Hum. Genet. 78, 947–960 (2006).
Zhu, M. et al. Mutations in the γ-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26). Am. J. Hum. Genet. 73, 1082–1091 (2003).
Yarmola, E.G., Somasundaram, T., Boring, T.A., Spector, I. & Bubb, M.R. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 275, 28120–28127 (2000).
Tartaglia, M. & Gelb, B.D. Noonan syndrome and related disorders: genetics and pathogenesis. Annu. Rev. Genomics Hum. Genet. 6, 45–68 (2005).
Ibrahimi, O.A., Zhang, F., Eliseenkova, A.V., Linhardt, R.J. & Mohammadi, M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum. Mol. Genet. 13, 69–78 (2004).
Lindhurst, M.J. et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 365, 611–619 (2011).
Kholmanskikh, S.S., Dobrin, J.S., Wynshaw-Boris, A., Letourneau, P.C. & Ross, M.E. Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J. Neurosci. 23, 8673–8681 (2003).
Kholmanskikh, S.S. et al. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat. Neurosci. 9, 50–57 (2006).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Cooper, G.M. et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods 7, 250–251 (2010).
Grantham, R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974).
Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Gilissen, C. et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 87, 418–423 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Acknowledgements
We thank the families for their contribution to this study. We thank all members of the Northwest Genomics Center and Genomic Disorders Group Nijmegen, as well as personnel from the Microarray Facility and Sequencing Facility Nijmegen, for excellent technical assistance. This work was supported by grants from the US National Institutes of Health (NS058721 to W.B.D. and NS048120 to M.E.R.), the Netherlands Organization for Health Research and Development (ZonMW; 917-66-363 and 911-08-025 to J.A.V.), the European Union (EU)-funded TECHGENE project (Health-F5-2009-223143 to J.A.V.) and the AnEUploidy project (LSHG-CT-2006-37627 to A.H., B.W.M.v.B., H.G.B. and J.A.V.). J.-B.R. is supported by a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research. T.R. is supported by an Australian National Health and Medical Research Council (NHMRC) postdoctoral fellowship. We thank the Simons Foundation Autism Research Initiative (SFARI) that provided control exome data (191889), the National Institute of Environmental Health Services (NIEHS) Environmental Genome Project for providing support for this project (HHSN273200800010C) and the NHLBI GO Exome Sequencing Project and its ongoing studies that produced and provided exome variant calls for comparison, including the Lung GO Sequencing Project (HL-102923), the Women's Health Initiative (WHI) Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Author information
Authors and Affiliations
Contributions
D.T.P., W.B.D., A.V. and H.G.B. designed the study. J.-B.R., B.W.M.v.B. and A.H. designed and performed the genetics experiments. S.S.K. performed the experiments in lymphoblastoid cell lines. J.-B.R., A.H., B.J.O. and C.G. performed the bioinformatics experiments. B.J.O. analyzed the control exome data sets. S.G. contributed to the genetics experiments. C.T.S. and S.L.C. prepared DNA samples and lymphoblastoid cell lines. O.A.A.-R., J.F.A., N.C., V.D.-G., A.E.F., J.-P.F., K.W.G., M.K., T.K., G.M.S.M., M.J.M.N., C.M.A.v.R.-A., T.R., B.B.A.d.V., M.M., V.M.S., A.V., H.G.B., D.T.P. and W.B.D. recruited and evaluated the study subjects. J.A.R. analyzed the Signature Genomic Laboratories data set. J.S. supervised B.J.O., J.A.V. supervised A.H., C.G. and S.G., H.G.B. supervised B.W.M.v.B., M.E.R. supervised and evaluated data with S.S.K. and wrote sections related to lymphoblastoid cell lines, and W.B.D. supervised J.-B.R. J.-B.R. and W.B.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–5 and Supplementary Tables 1–6 (PDF 640 kb)
Rights and permissions
About this article
Cite this article
Rivière, JB., van Bon, B., Hoischen, A. et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet 44, 440–444 (2012). https://doi.org/10.1038/ng.1091
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1091
This article is cited by
-
Obsessive–compulsive symptoms in ACTG1-associated Baraitser-Winter cerebrofrontofacial syndrome
Journal of Neural Transmission (2022)
-
Human deafness-associated variants alter the dynamics of key molecules in hair cell stereocilia F-actin cores
Human Genetics (2022)
-
Confirmation of FZD5 implication in a cohort of 50 patients with ocular coloboma
European Journal of Human Genetics (2021)
-
Okur-Chung neurodevelopmental syndrome-linked CK2α variants have reduced kinase activity
Human Genetics (2021)
-
Impaired eIF5A function causes a Mendelian disorder that is partially rescued in model systems by spermidine
Nature Communications (2021)