Abstract
The acronym VATER/VACTERL association describes the combination of at least three of the following congenital anomalies: vertebral defects (V), anorectal malformations (A), cardiac defects (C), tracheoesophageal fistula with or without esophageal atresia (TE), renal malformations (R), and limb defects (L). We aimed to identify highly penetrant de novo copy number variations (CNVs) that contribute to VATER/VACTERL association. Array-based molecular karyotyping was performed in a cohort of 41 patients with VATER/VACTERL association and 6 patients with VATER/VACTERL-like phenotype including all of the patients’ parents. Three de novo CNVs were identified involving chromosomal regions 1q41, 2q37.3, and 8q24.3 comprising one (SPATA17), two (CAPN10, GPR35), and three (EPPK1, PLEC, PARP10) genes, respectively. Pre-existing data from the literature prompted us to choose GPR35 and EPPK1 for mouse expression studies. Based on these studies, we prioritized GPR35 for sequencing analysis in an extended cohort of 192 patients with VATER/VACTERL association and VATER/VACTERL-like phenotype. Although no disease-causing mutation was identified, our mouse expression studies suggest GPR35 to be involved in the development of the VATER/VACTERL phenotype. Follow-up of GPR35 and the other genes comprising the identified duplications is warranted.
Similar content being viewed by others
Introduction
The VATER/VACTERL association (OMIM #192350) refers to the rare, non-random co-occurrence of the following component features (CFs): vertebral defects (V), anorectal malformations (A), cardiac defects (C), tracheoesophageal fistula with or without esophageal atresia (TE), renal malformations (R), and limb defects (L).1 Population-based epidemiological studies have reported a prevalence among infants of 1 in 10 000 to 1 in 40 000 with male predominance.2, 3, 4 Since this multisystem malformation complex has been associated with reduced reproduction, it is reasonable to assume that a significant proportion of cases are caused by de novo mutations. In accordance with this, previous studies have identified several de novo chromosomal microaberrations, which are likely to be disease causing.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 However, none of these de novo chromosomal microaberrations has led to the identification of a disease-causing gene and the etiology in most cases is still unknown.
Using a SNP-array-based genome-wide approach, we systematically screened 41 patients with VATER/VACTERL association and 6 patients with a VATER/VACTERL-like phenotype, including all of the patients’ parents, in order to identify causative de novo copy number variations (CNVs). Pre-existing data from the literature were used to choose candidate genes within the identified CNVs for in situ hybridization (ISH) on sections from mouse embryos at embryonic days (E) 12.5−14.5. Based on the ISH, candidate genes were prioritized for sequencing analysis in an extended cohort of 192 patients with VATER/VACTERL association and VATER/VACTERL-like phenotype in order to identify further disease-causing mutations.
Materials and methods
Subjects and DNA isolation
This study was approved by the local ethics committee and all families provided written informed consent. Families were contacted through the German self-help organization for patients with anorectal malformations (SoMA e.V.) and various pediatric surgical departments throughout Germany. Among the patients was one aborted female fetus, enrolled through the Department of Paidopathology, University of Bonn. All patients and their families were personally recruited by one of four experienced physicians starting in 2009. None of the patients used for array-based analysis has been reported in our previous studies.15, 16 To optimize for detection of de novo CNVs, we only included patients with a negative family history of the disease. Forty-one patients presented with three or more CFs of the VATER/VACTERL association, while six patients presented with only two, with or without further anomalies (VATER/VACTERL-like phenotype).1 For the mutation-sequencing analysis, an additional 59 patients with VATER/VACTERL association and 60 patients with VATER/-VACTERL-like phenotype recruited within Germany were added to the patient sample, of which 18 patients were previously described in Schramm et al.15, 16 An additional 26 patients with VATER/VACTERL association were recruited within the AGORA project and were added to the sample for the mutation-sequencing analysis. The AGORA project is run within the Netherlands and patients are being recruited by pediatric surgeons and clinical geneticists of the Radboud University, Nijmegen Medical Centre, Nijmegen, the Netherlands. The complete sample for mutation-sequencing analysis comprised 192 patients (n=132 patients with VATER/VACTERL association; n=60 patients with VATER/-VACTERL-like phenotype). As all of the recruited patients in Germany and the Netherlands were part of research projects on the genetics of anorectal malformations, all patients had anorectal malformations. Blood or saliva was taken from the patient and from both parents for DNA isolation of genomic DNA, carried out using the Chemagic Magnetic Separation Module I (Chemagen, Baesweiler, Germany) or, in case of saliva samples, the Oragene DNA Kit (DNA Genotek Inc., Kanata, ON, Canada).
Array genomic hybridization analysis
For molecular karyotyping, we used the Illumina HumanOmni1-Quad-v1 BeadChip (marker content, 1 140 419; median marker spacing 1.5 kb) (San Diego, CA, USA). A DNA sample was considered to have failed if less than 98% of the markers were generated on the respective BeadChip.
Paternity testing
Paternity testing was performed using the GenomeStudio (v2010.3, www.illumina.com/) genotyping module in accordance with the manufacturer’s recommendations.
CNV analysis
To identify potential CNVs, the SNP fluorescence intensity data from all patients and their parents were analyzed with QuantiSNP (v2.1 and v2.2, www.well.ox.ac.uk/QuantiSNP/) using an Objective Bayes Hidden-Markov model for calling putative CNVs.17 CNVs with a log Bayes factor below 7 were discarded.18 The results were also checked visually using the GenomeStudio genotyping module (v2010.3, www.illumina.com/) for marker and signal quality and the occurrence of mosaic aberrations. The Cartagenia Bench software (Cartagenia n.v., Leuven, Belgium) was used to filter detected CNVs for de novo events, variable regions in our control cohort of 531 unaffected controls, and gene content. The remaining aberrations were checked for gene content according to UCSC human genome browser assembly build 18, and for their presence in the databases DGV (http://dgvbeta.tcag.ca/dgv/app/home?ref=NCBI36/hg18) and DECIPHER (http://decipher.sanger.ac.uk/). CNVs identified to exist in either of these databases, with at least 10 full overlapping reports in DGV and at least five full overlapping reports in DECIPHER, were excluded. However, we did not exclude CNVs although they were fully overlapping by at least five reports in DECIPHER, if one of the associated phenotypes comprised any of the classical component features of the VATER/VACTERL association: e.g. esophageal atresia (TE), anorectal malformation (A), cardiac defects (C).
Quantitative PCR
For verification of the filtered CNVs, qPCR was performed as described previously using SYBR Green for detection.19 All primer sequences are listed in Supplementary Table 2.
In situ hybridization of mouse embryo sections
To prioritize candidate genes within the remaining CNVs for further mutation analysis in larger patient cohorts, ISH on sections from mouse embryos at E12.5–14.5, the crucial embryonic timeframe for the VATER/VACTERL association-relevant tissues, was performed as previously described.20 Antisense probes were generated by PCR from a total cDNA library from E10.5 where PCR products contained a 3′ T7 RNA polymerase-binding site for in vitro transcription. Probes were purified using G-50 Sephadex columns (GE Healthcare, Munich, Germany). The 903-bp probe for Gpr35 (starting 13 bp downstream of the ATG start site) spans nearly the entire 924-bp coding region and recognizes both known mRNA variants. Eppk1 is encoded by a single, large exon (>20 kb) consisting of 16 plakin repeat domains, which are the hallmark of all plakin family members.21 Thus, the unique 647-bp probe for Eppk1 spans the 3′UTR. Detection of alkaline phosphatase activity was visualized using BM Purple (Roche Diagnostics, Mannheim, Germany). Following staining, slides were quickly dehydrated in 80% and then 100% ethanol, cleared twice for 1 min in xylene (Roth, Karlsruhe, Germany) and coverslips were mounted with Entellan mounting medium (Merck, Darmstadt, Germany). Photographs were obtained using AxioVision software (Zeiss, Göttingen, Germany) with a Zeiss AxioCam and SteREO DiscoveryV12 microscope. Three sections from at least two different embryos were analyzed for each stage shown and representative images are provided.
Sequence analysis
Analysis of the protein-coding exons 5 and 6 of the human GPR35 gene was performed using polymerase chain reaction (PCR), and PCR-amplified DNA products were subjected to direct automated sequencing (3130XL Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s specifications. Both strands of each amplicon were sequenced. All primer sequences are available upon request.
Results
Array analysis
Molecular karyotyping identified five possible de novo aberrations with gene content in five of the 47 patients. Three of these were confirmed as being de novo by qPCR (Table 1, Supplementary Tables 1 and 2, and Supplementary Figure), the others were found to be inherited by one parent (see Supplementary Tables 1 and 2). In the three confirmed cases, there were no other dysmorphic features, congenital anomalies (including structural brain anomalies or evidence of neurologic impairment), or other medical issues except those noted below.
Case 1 is a male patient with butterfly vertebra (V), anal atresia without fistula (A), atrial and ventricular septal defects (C), esophageal atresia IIIb (according to Vogt)22 (TE), bilateral dystopic kidneys, and left-sided renal dysplasia (R). A de novo microduplication was found at 1q41 (Table 1) with an estimated size of 132 kb, comprising exons 6–9 of SPATA17, encoding the spermatogenesis-associated protein 17.
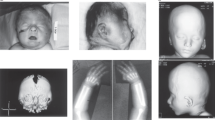
Case 2 is an aborted female fetus. The pregnancy was terminated at 30+1 weeks of gestation because of sonographically proven severe bilateral renal dysplasia. Macroscopic and histological findings obtained at autopsy revealed a cloacal malformation with anal atresia (A), bilateral renal dysplasia (R), urethral agenesis, a secondary bell-shaped thorax, and Potter-sequence facies due to anhydramnion (Figure 1). A de novo microduplication was found at 2q37.3 (Table 1) with an estimated size of 25 kb, comprising exons 3–4 of one splice variant of Calpain-10 (CAPN10) and exons 3–6 of GPR35, encoding the G-protein-coupled receptor 35.
Case 3 is a male patient with butterfly vertebra (V), anal atresia with recto-vesical fistula (A), congenital subvalvular aortic stenosis and ventricular septal defect (C), esophageal atresia IIIb (according to Vogt)22 (TE), high-grade bilateral vesico-ureteral reflux (III° and IV°) (R), and bilateral cryptorchidism. A de novo microduplication was found at 8q24.3 (Table 1) with an estimated size of 120 kb. This region comprises parts of exon 1 of the epiplakin gene (EPPK1), the complete plectin-1 gene (PLEC-1), and exons 4–11 of the gene coding for poly(ADP-ribose)polymerase 10 (PARP10).
CNVs found in DGV with partial overlap of the three duplicated regions of interest (1q41, 2q37.3, 8q24.3) are listed in Supplementary Table 3. In DECIPHER, no partially but only fully overlapping CNVs were found. Interestingly, the duplicated region 8q24.3 found in case 3 contains partially overlapping CNVs more frequently than the other duplications, most likely due to the adjacent repeat elements mapping to the boundaries of this region.
Gene prioritization and ISH analysis
A database search on expression data and targeted deletions in mice for the genes Spata17, Capn10, Gpr35, Eppk1, Plec1, and Parp10 using the Mouse Genome Informatics (MGI) database was performed to narrow down the candidate gene list. Entries provide expression data from the Eurexpress project23 freely available from the Jackson Laboratory Mouse Genome Informatics database (http://www.informatics.jax.org/). As expression of Spata17, which encodes a protein related to apoptosis of spermatogenic cells, is restricted to the testis in several species,24, 25 it was not given high priority for follow-up. The expression of Capn10 coding for a member of the calpain-like cysteine protease family is moderate and ubiquitous at E14.5. Targeted deletion of Capn10 resulted in viable mice showing increased adiposity and metabolic defects26 in the homozygous state. Hence, it was not given high priority. Plectin-1 interlinks intermediate filaments with microtubules and microfilaments, and MGI expression data on Plec1 during development were limited to the retina after E14.0, but are likely to be elsewhere also, considering homozygous knockout embryos exhibiting neonatal death with skin blistering and defects in integument.27 Mutations in PLEC1 in human have been found to be associated with the epidermolysis bullosa phenotypic spectrum.28 No expression data were found for Parp10, but this gene shares some common exons with Plec1.29 In their studies of targeted deletion of Plec1, Fuchs et al,29 as a consequence of exon sharing between Plec1 and Parp10, generate a P1c−/− mouse line that represents in fact a double knockout (P1c−/−/Parp10−/−). Although this targeted homozygous deletion was found to affect the nervous system and the motor nerve conduction velocity of the mice model investigated, it can be ruled out that this was due to the targeted deletion of Parp10, as the conditional nes-Cre/plecf/Δ mouse model (where plectin was knocked down, leaving Parp10 unaffected) showed comparable phenotypes.29 Owing to the mouse expression data in MGI and lack of congenital malformations in both homozygous murine knockout models of Plec1 and Parp10, both genes were not given high priority for follow-up.
The most promising candidates from the MGI expression data were found to be Gpr35 and Eppk1. GPR35 is a member of the G-protein-coupled receptor superfamily of transmembrane proteins involved in a variety of physiological pathways. Previous studies showed expression of GPR35 in tissues of the gastrointestinal tract, nervous system, and lymphoid tissues. It was reported that kynurenic acid might be an endogenous ligand for GPR35,30 but more recent data suggested lysophosphatidic acid (specifically 2-oleoyl LPA) to be a more likely candidate.31 As Oka et al31 suggested that RhoA signaling might be activated downstream of GPR35 (in vitro) and RhoA is involved in the non-canonical planar cell polarity (PCP) pathway, it is tempting to speculate that one role of GPR35 might be cell motility/polarity during embryonic development. According to MGI, Gpr35 expression has been detected at E14.5 within the stomach, hindgut, rectum, midgut, bladder fundus region, and urethra. Eppk1 was expressed at E14.5 in the epidermis, lung, and alimentary canal tissues, including the stomach, esophagus, and gut. However, complete deletion of Eppk1 results in normal embryonic development,32 whereas there is currently no model for targeted deletion of Gpr35 in mice. According to these pre-existing data, we decided to validate the expression studies on Gpr35 and Eppk1 between E12.5–14.5, which is the equivalent of human gestational weeks 6–8,33 encompassing the relevant stages of development in mice for organ systems involved in the etiology of VATER/VACTERL.
The expression of Eppk1 at E12.5 was found within the liver, oral epithelium, nasal passages, brain, and neural tube. At E14.5, it was additionally found in the developing tooth buds and esophageal epithelium (Figure 2). Expression of Gpr35 at E12.5 was found in the mesonephros, the heart, the esophagus and trachea, the dorsal root ganglia, the genital tubercle, and the rectal region, and a similar expression pattern was detected at E14.5 (Figure 2). This expression not only reflected the affected organ systems of the patient/fetus exhibiting the GPR35 comprising duplication but also showed expression in tissues relevant to the VATER/VACTERL association in general. Although the genital tubercle and the dorsal root ganglia do not primarily belong to the tissues reflected by the CFs of the VATER/VACTERL association, we point them out because a significant proportion of patients present with genital anomalies/defects arising from the genital tubercle.3 The neurons of the dorsal root ganglia are closely affiliated with vertebral development and are derived from the neural crest cells, which are involved in the formation of tissues including the cardiac outflow tract and enteric nervous system of the gut, suggesting a role in VATER/VACTERL association.
Embryonic mouse expression of Eppk1 and Gpr35. (a) Eppk1 expression at E12.5 is detectable in the forebrain (fb), midbrain (mb), inner ear (ie), liver (li), nasal passage (np), genital tubercle (gt), dorsal root ganglia (drg), and spinal cord (sc). (b) Eppk1 expression at E14.5 is detectable in the liver, Rathke’s pouch (Rp), esophagus (e), corpus striatum mediale (csm), genital tubercle, tooth bud (tb), spinal cord, and nasal passages. (c) Gpr35 expression at E12.5. Transcripts are evident in the liver, trigeminal ganglion (trg), vestibulocochlear ganglion (vcg), heart ventricle (h), stomach (st), mesonephros (me), genital tubercle, dorsal root ganglia, and surrounding the maxillo-mandibular cleft (mmc). (d) Gpr35 expression at E14.5 is detectable in the liver, corpus striatum mediale, hypothalamus (ht), tooth bud, lung (lu), genital tubercle, and rectum.
Candidate gene analysis
We further prioritized Gpr35 over Eppk1 as a result of both its expression and available information in the literature, and carried out mutation analysis of GPR35 in 192 VATER/VACTERL/VACTERL-like patients. Sanger sequencing of the two protein-coding exons (exon 5 and 6) of GPR35 in these 192 patients (including case 2) did not identify any sequence variant of likely pathological significance. Heterozygous sequence variants with a minor allele frequency less than 1% (rs139197368, rs35146537, rs201441371) were also checked for their de novo status. They were found to be inherited by one parent, with the exception of one sequence variant, where the father’s DNA was not available for testing. Pathogenicity prediction with four algorithms (PolyPhen-2, SNPs&Go, MutPred, SIFT) predicted none of the three SNPs unanimously as disease causing. Two of these variants have been predicted to be benign in all four prediction algorithms applied. SNP rs201441371 leading to a p.Val237Met substitution was classified as possibly disease causing in two and benign in the other two pathogenicity-prediction algorithms. Hence, incomplete penetrance cannot be excluded for this variant and should warrant functional studies.
Discussion
Although only a few large studies have been published, there are several lines of evidence that de novo CNVs are associated with the expression of structural birth defects.34 Depending on the investigated birth defect, de novo CNVs have been assumed to be causative in up to 20% of cases.35 In our study of patients with VATER/VACTERL association and VATER/VACTERL-like phenotype, we identified three (6%) likely causative de novo CNVs at 1q41, 2q37.3, and 8q24.3. These CNVs contained between one and three genes, including GPR35 and EPPK1, which are known, according to expression data from MGI, to participate in the development of tissues relevant to the VATER/VACTERL-like phenotype (Table 1). For candidate gene mutation analysis, we prioritized GPR35 based on its expression in mice in the rectal region, the genital tubercle, and the mesonephros, showing a clear overlap with the malformed tissues in the duplication-carrying patient (case 2) (Figures 1 and 2). Sequence analysis of the protein-coding exons did not identify any disease-causing mutation. It is therefore unlikely that mutations affecting the GPR35 gene are a frequent cause of VATER/VACTERL association; however, our patient cohort might have been too small to detect rare causal mutational events. Furthermore, we may have missed mutations by not analyzing the non-protein-coding exons or the promoter region. Yet, according to our mouse expression data reflecting both the affected organ systems of the patient/fetus exhibiting the GPR35 comprising duplication and the tissues relevant to the VATER/VACTERL association, GPR35 seems to be a promising candidate gene being worth of further follow-up in genetic as well as functional studies. Nevertheless, as we cannot exclude EPPK1, SPATA17, CAPN10, PLEC1, or PARP10 as candidate genes, follow-up of these genes is also warranted.
Our study did not investigate whether the identified microduplications alter the expression of the genes involved or lead to altered gene products. This will require further studies. Particularly, in the case of a negative effect on the gene function, there remains also the possibility that the anomalies seen in patients are the results of autosomal-recessive mutations, with one allele being altered by the de novo event and the second allele being altered by an inherited change. We did not exhaustively investigate this possibility. However, the lack of a mutation in GPR35 in case 2 and the lack of GPR35 mutations in the extended patients sample render this possibility unlikely for GPR35. We can also not exclude the possibility that the expression of a gene residing in a region flanking a microduplication could be affected by means of a position effect. However, thorough screening of the flanking regions failed to detect obvious candidate genes. Finally, it remains possible that there were disease-associated CNVs among the many inherited CNVs and that the full penetrance may only be exerted by interaction with an environmental insult, as has been recently demonstrated for hypoxia and fibroblast growth factor signaling in the etiology of congenital scoliosis.36
In summary, a genome-wide analysis in patients with VATER/VACTERL association or VATER/VACTERL-like phenotype has identified three potentially pathogenic CNVs affecting genes involved in the development of the phenotype-relevant tissues. Although mutation screening of the most promising candidate gene, GPR35, did not confirm it as a frequent cause of the disorder, new insights were gained and the identified variant loci warrant further evaluation.
References
Quan L, Smith DW : The VATER association. Vertebral defects, anal atresia, T-E fistula with esophageal atresia, radial and renal dysplasia: a spectrum of associated defects. J Pediatr 1973; 82: 104–107.
Khoury MJ, Cordero JF, Greenberg F, James LM, Erickson JD et al: A population study of the VACTERL association: evidence for its etiologic heterogeneity. Pediatrics 1983; 71: 815–820.
Czeizel A, Ludányi I : An aetiological study of the VACTERL-association. Eur J Pediatr 1985; 144: 331–337.
Botto LD, Khoury MJ, Mastroiacovo P, Castilla EE, Moore CA, Skjaerven R et al: The spectrum of congenital anomalies of the VATER association: an international study. Am J Med Genet 1997; 71: 8–15.
McNeal RM, Skoglund RR, Francke U : Congenital anomalies including the VATER association in a patient with del(6)q deletion. J Pediatr 1977; 91: 957–960.
Aynaci FM, Celep F, Karaguzel A, Baki A, Yildiran A : A case of VATER association associated with 9qh+. Genet Couns 1996; 7: 321–322.
Walsh LE, Vance GH, Weaver DD : Distal 13q deletion syndrome and the VACTERL association: case report, literature review, and possible implications. Am J Med Genet 2001; 98: 137–144.
Cinti R, Priolo M, Lerone M, Gimelli G, Seri M, Silengo M et al: Molecular characterisation of a supernumerary ring chromosome in a patient with VATER association. J Med Genet 2001; 38: E6.
Prieto JC, Garcia NM, Elder FF, Zinn AR, Baker LA : Phenotypic expansion of the supernumerary derivative (22) chromosome syndrome: VACTERL and Hirschsprung’s disease. J Pediatr Surg 2007; 42: 1928–1932.
Yamada K, Uchiyama A, Arai M, Kubodera K, Yamamoto Y, Orii KO et al: Severe upper airway stenosis in a boy with partial monosomy 16p13.3pter and partial trisomy 16q22qter. Congenit Anom (Kyoto) 2009; 49: 85–88.
De Jong EM, Douben H, Eussen BH, Felix JF, Wessels MW, Poddighe PJ et al: 5q11.2 deletion in a patient with tracheal agenesis. Eur J Hum Genet 2010; 18: 1265–1268.
Van der Veken LT, Dieleman MMJ, Douben H, van de Brug JC, van de Graaf R, Hoogeboom AJ et al: Low grade mosaic for a complex supernumerary ring chromosome 18 in an adult patient with multiple congenital anomalies. Mol Cytogenet 2009; 3: 13.
Zen PRG, Riegel M, Rosa RF, Pinto LL, Graziadio C, Schwartz IV et al: Esophageal stenosis in a child presenting a de novo 7q terminal deletion. Eur J Med Genet 2010; 53: 333–336.
Solomon BD, Pineda-Alvarez DE, Hadley DW, Keaton AA, Agochukwu NB, Raam MS et al: De novo deletion of chromosome 20q13.33 in a patient with tracheo-esophageal fistula, cardiac defects, and genitourinary anomalies implicates GTPBP5 as a candidate gene. Birth Defects Res A Clin Mol Teratol 2011b; 91: 862–865.
Schramm C, Draaken M, Bartels E, Boemers TM, Aretz S, Brockschmidt FF et al: De novo microduplication at 22q11.21 in a patient with VACTERL association. Eur J Med Genet 2011a; 54: 9–13.
Schramm C, Draaken M, Bartels E, Boemers TM, Schmiedeke E, Grasshoff-Derr S et al: De novo duplication of 18p11.21-18q12.1 in a female with anorectal malformation. Am J Med Genet A. 2011b; 155A: 445–449.
Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P et al: An objective Bayes Hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007; 35: 2013–2025.
Engels H, Wohlleber E, Zink A, Hoyer J, Ludwig KU, Brockschmidt FF et al: A novel microdeletion syndrome involving 5q14.3-q15: clinical and molecular cytogenetic characterization of three patients. Eur J Hum Genet 2009; 17: 1592–1599.
Draaken M, Giesen CA, Kesselheim AL, Jabs R, Aretz S, Kugaudo M et al: Maternal de novo triple mosaicism for two single OCRL nucleotide substitutions (c.1736A>T,c.1736A>G) in a Lowe syndrome family. Hum Genet 2011; 129: 513–519.
Pennimpede T, Proske J, König A, Vidigal JA, Morkel M, Bramsen JB et al: In vivo knockdown of Brachyury results in skeletal defects and urorectal malformations resembling caudal regression syndrome. Dev Biol 2012; 372: 55–67.
Spazierer D, Fuchs P, Pröll V, Janda L, Oehler S, Fischer I et al: Epiplakin gene analysis in mouse reveals a single exon encoding a 725-kDa protein with expression restricted to epithelial tissues. J Biol Chem 2003; 278: 31657–31666.
Vogt EC : Congenital esophageal atresia. Am J Roentgenol 1929; 22: 463–465.
Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D et al: A high-resolution anatomical atlas of the transcriptome in the mouse embry. PLoS Biol 2011; 9: e1000582.
Deng Y, Hu LS, Lu GX : Expression and identification of a novel apoptosis gene Spata17 (MSRG-11) in mouse spermatogenic cells. Acta Biochim Biophys Sin (Shanghai) 2006; 38: 37–45.
Nie D, Liu Y, Xiang Y : Overexpression a novel zebra fish spermatogenesis-associated gene 17 (SPATA17) induces apoptosis in GC-1 cells. Mol Biol Rep 2011; 38: 3945–3952.
Cheverud JM, Fawcett GL, Jarvis JP, Norgard EA, Pavlicev M, Pletscher LS et al: Calpain-10 is a component of the obesity-related quantitative trait locus Adip1. J Lipid Res 2010; 51: 907–913.
Andrä K, Lassmann H, Bittner R, Shorny S, Fässler R, Propst F et al: Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev 1997; 11: 3143–3156.
Pfendner E, Rouan F, Uitto J : Progress in epidermolysis bullosa: the phenotypic spectrum of plectin mutations. Exp Dermatol 2005; 14: 241–249.
Fuchs P, Zörer M, Reipert S, Rezniczek GA, Propst F, Walko G et al: Targeted inactivation of a developmentally regulated neural plectin isoform (plectin 1c) in mice leads to reduced motor nerve conduction velocity. J Biol Chem 2009; 284: 26502–26509.
Wang J, Simonavicius N, Wu X, Swaminath G, Reagan J, Tian H et al: Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem 2006; 281: 22021–22028.
Oka S, Ota R, Shima M, Yamashita A, Sugiura T : GPR35 is a novel lysophosphatidic acid receptor. Biochem Biophys Res Commun 2010; 395: 232–237.
Goto M, Sumiyoshi H, Sakai T, Fässler R, Ohashi S, Adachi E et al: Elimination of epiplakin by gene targeting results in acceleration of keratinocyte migration in mice. Mol Cell Biol 2006; 26: 548–558.
O’Rahilly R : Early human development and the chief sources of information on staged human embryos. Eur J Obstet Gynecol Reprod Biol 1979; 9: 273–280.
Southard AE, Edelmann LJ, Gelb BD : Role of copy number variants in structural birth defects. Pediatrics 2012; 129: 755–763.
Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS et al: Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis 2011; 6: 592–602.
Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O'Reilly VC et al: A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 2012; 149: 295–306.
Acknowledgements
We thank all patients and their parents for their participation in this study. We thank the German self-help organization for providing people with anorectal malformations (SoMA e.V.). Grants were sponsored by the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF; grant no. 01GM08107) and The BONFOR program of the University of Bonn (grant no. O-149.0093 to CS, grant no. O-149.0099 to EB, and grant no. O-149.0096 to GCD). TP, LW, EB, ES, SG-D, SM, SH, SH-C, BGH, MMN, ML, HR, and MD are members of the ‘Network for the Systematic Investigation of the Molecular Causes, Clinical Implications and Psychosocial Outcome of Congenital Uro-Rectal Malformations (CURE-Net)’, which is supported by a research grant from the BMBF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Hilger, A., Schramm, C., Pennimpede, T. et al. De novo microduplications at 1q41, 2q37.3, and 8q24.3 in patients with VATER/VACTERL association. Eur J Hum Genet 21, 1377–1382 (2013). https://doi.org/10.1038/ejhg.2013.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.58
Keywords
This article is cited by
-
Genome-wide identification of disease-causing copy number variations in 450 individuals with anorectal malformations
European Journal of Human Genetics (2023)
-
A review of genetic factors contributing to the etiopathogenesis of anorectal malformations
Pediatric Surgery International (2018)
-
Underlying genetic factors of the VATER/VACTERL association with special emphasis on the “Renal” phenotype
Pediatric Nephrology (2016)
-
Copy number variations in 375 patients with oesophageal atresia and/or tracheoesophageal fistula
European Journal of Human Genetics (2016)
-
PCSK5 mutation in a patient with the VACTERL association
BMC Research Notes (2015)