Abstract
Dent’s disease is an X-linked renal tubulopathy caused by mutations mainly affecting the CLCN5 gene. Defects in the OCRL gene, which is usually mutated in patients with Lowe syndrome, have been shown to lead to a Dent-like phenotype called Dent disease 2. However, about 20% of patients with Dent’s disease carry no CLCN5/OCRL mutations. The disease’s genetic heterogeneity is accompanied by interfamilial and intrafamilial phenotypic heterogeneity. We report on a case of Dent’s disease with a very unusual phenotype (dysmorphic features, ocular abnormalities, growth delay, rickets, mild mental retardation) in which a digenic inheritance was discovered. Two different, novel disease-causing mutations were detected, both inherited from the patient’s healthy mother, that is a truncating mutation in the CLCN5 gene (A249fs*20) and a donor splice-site alteration in the OCRL gene (c.388+3A>G). The mRNA analysis of the patient’s leukocytes revealed an aberrantly spliced OCRL mRNA caused by in-frame exon 6 skipping, leading to a shorter protein, but keeping intact the central inositol 5-phosphatase domain and the C-terminal side of the ASH-RhoGAP domain. Only wild-type mRNA was observed in the mother’s leukocytes due to a completely skewed X inactivation. Our results are the first to reveal the effect of an epistatic second modifier in Dent’s disease too, which can modulate its expressivity. We surmise that the severe Dent disease 2 phenotype of our patient might be due to an addictive interaction of the mutations at two different genes.
Similar content being viewed by others
Introduction
Dent’s disease is now the generally accepted name for a group of X-linked renal tubular disorders, including X-linked recessive nephrolithiasis with renal failure, X-linked recessive hypophosphatemic rickets, and idiopathic low-molecular weight (LMW) proteinuria.1, 2, 3
Dent’s disease is caused mainly by mutations in the CLCN5 gene (Dent disease 1, MIM #300009) located on chromosome Xp11.22, which encodes for the 746 amino-acid ClC-5 chloride channel implicated in the tubular endocytotic reabsorption of albumin and LMW proteins. ClC-5 was first thought to provide a shunt conductance in early endosomes, enabling efficient intraluminal acidification by V-type H+ ATPase.4, 5, 6 It has recently been demonstrated, however, that ClC-5 functions as a Cl−/H+ antiporter when activated by positive voltages.7, 8
No CLCN5 gene mutations are detected in approximately 40% of patients with the classic symptoms of Dent’s disease, suggesting a locus heterogeneity. The OCRL gene located on chromosome Xq26.1, whose mutations cause Lowe syndrome, has recently been found altered in 20% Dent’s patients,9 but about 20% of patients carry neither CLCN5 nor OCRL mutations.10, 11, 12, 13
Dent’s disease tends to become manifest in childhood or early adult life. It is characterized by LMW proteinuria, hypercalciuria, medullary nephrocalcinosis, nephrolithiasis, other tubular dysfunctions, and renal failure in various combinations. Different groups of researchers have recently reported on case series of patients with atypical or rare Dent disease 1 phenotypic signs, such as episodic night blindness,14 Bartter-like syndrome,15, 16 growth hormone deficiency,17 and proteinuria with histological evidence of focal segmental glomerulosclerosis.18, 19
Dent’s disease patients carrying OCRL gene mutations (Dent disease 2, MIM#300555) have none of the classic symptoms accompanying renal tubulopathy in Lowe syndrome, that is mental retardation, bone disease, growth retardation, congenital cataracts, delayed motor milestones. This milder phenotype is not attributable to less severe changes in protein expression or enzyme activity, as both are significantly reduced or absent.10, 11, 12, 13 The renal symptoms of Lowe syndrome are very similar to those of Dent’s disease, though the characteristics of the patients’ tubular dysfunction may differ. To date, there are reports of around 45 patients with Dent disease 2; a minority of them have revealed a few mild extrarenal signs, such as mild intellectual impairment, ocular abnormalities, and short stature.10 Recently however, two different OCRL mutations each causing both Dent disease 2 and Lowe syndrome even in the same family have been described.20 In short, Dent’s disease seems to be characterized by a genetic and phenotypic heterogeneity.
Here, we describe and discuss an atypical case involving a syndromic variant of Dent’s disease, with documented CLCN5 and OCRL mutations.
Materials and methods
Case report
A second-born, 6-year-old boy was referred for tubulopathy, rickets, and syndromic features including microcephaly and dysmorphic facies, that is a triangular face, severe prognathism, anterior crossbite, hypomaxillia, malocclusion, large, low-set ears, low forehead, and arched palate (Figures 1a and b).
Front and lateral view of the patient at the age of 6 years. (a, b) Note triangular face, severe prognathism, hypomaxillia, anterior crossbite, large low-set ears, low forehead. (c) Note rickets, bilateral and symmetrical widening of the metaphyseal regions, curvature of both the femoral, and the tibial diaphyses.
In his family history he had a paternal grandmother with bilateral tibia vara, a first-degree cousin with mental retardation of unknown origin, and a maternal grandfather who had suffered from nephrolithiasis since his youth.
The patient’s medical history included orthopedic investigations at 3 years of age for tibia vara due to rickets and neurological investigations for attention deficit hyperactivity disorder. MRI showed asymmetry of the brain hemispheres and ventricular system, with septum pellucidum and right ventricle deviation, and partial interdigitation of the medial frontal cerebral cortex. The boy had undergone genetic analysis for mutations in the FRAXA and FRAXE genes, and for copy number variations by CGH arrays, the results of which were negative.
At the time of our investigations, the patient had:
-
normal renal function, electrolytemia, parathyroid hormone levels, and blood pH; nonselective aminoaciduria, hypophosphoremia (2.9–3.6 mg/dl) with low tubular phosphorus reabsorption (TRP 71–84%), hypercalciuria (UCa/UCr >0.3), LMW proteinuria (β2 microglobulin >90 000 μg/l), paroxysmal microhematuria, a bone age of 4 years and 3 months (according to Greulich and Pick criteria), high bone alkaline phosphatase (911 IU/l);
-
poor parenchymal differentiation and hyperechoic spots compatible with his hypercalciuria at renal ultrasound;
-
florid rickets with a radiologically evident reduction in bone calcium content, bilateral and symmetrical widening of the metaphyseal regions, curvature of both the femoral, and the tibial diaphyses at X-ray (Figure 1c);
-
peripapillary optic nerve atrophy with gray papillae (no other ocular alterations at slit lamp examination);
-
mild mental retardation (IQ 70), attention deficit hyperactivity disorder on WISC/R test.
Renal findings were suggestive of Dent’s disease, so genetic analyses were performed.
Molecular studies
DNA analysis
Genomic DNA from peripheral blood was extracted using NucleoSpin Blood Quick Pure minicolumns (Macherey-Nagel GmbH & Co. KG, Duren, Germany). Primers and PCR conditions for amplifying the coding region, intron–exon boundaries, and 5′UTR exons of the CLCN5 gene have been described elsewhere.21
The PCR products were analyzed using the Agilent bioanalyzer (Agilent Technologies, Waldbronn, Germany) and purified with the QIAquick DNA purification kit (Qiagen GmbH, Hilden, Germany). Direct automated PCR product sequencing was done with the ABI PRISM GENESCAN 373A DNA sequencer and the BigDye Terminator v1.1 Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA). The nomenclature of the mutations is based on the ClC-5 cDNA sequence NM_000084.2.
Sequences of all 23 coding exons and the flanking intronic sequence of the human OCRL gene were analyzed according to Addis et al.22 The nomenclature of the mutations is based on the cDNA sequence NM_000276.3.
All the nucleotide variations observed were confirmed by means of independent PCR reactions.
OCRL mRNA analysis
Total RNA was extracted from the patient’s and his mother’s leukocytes using Trizol reagent (Life Technologies, Monza, Italy) according to the manufacturer’s instructions, and 0.2 μg of total RNA was reverse transcribed using the TaqMan multiscribe RT-PCR system (PE Applied Biosystems). Five microlitre of cDNA were amplified using a forward primer located in exon 4 (5′-GTTCAAGAAGCAGAAGAAACTCT-3′) and a reverse primer located in exon 7 (5′-GTCTTCAAATCCAAGAAGCC-3′), and PCR conditions according to Addis et al.21 PCR products were purified and directly sequenced on ABI 3130XL apparatus (PE Applied Biosystems).
X-inactivation study
The androgen receptor (AR) X chromosome inactivation (XCI) study was done using a PCR method described elsewhere.23 The XCI pattern was classified as random (ratios 50:50; <80:20), skewed (≥80:20), or extremely skewed (≥95:5).
Results and discussion
The boy, whose phenotype was characterized by classical Dent’s disease tubulopathy (LMW proteinuria, hypercalciuria, and microlithiasis), was first tested for CLCN5 mutations and a frameshift truncating variant in exon 7 of the CLCN5 gene, the 992_995 ins CAGC (A249fs*20) mutation, was identified. This mutation had never been described before and was inherited from his healthy mother.
The patient’s DNA had already been analyzed for any FRAXA and FRAXE mutations, or copy number variations, using CGH arrays because of his multisystemic signs and symptoms, but no abnormalities had come to light. The phenotype was highly unusual for Dent disease 1, however, and more suggestive of a mild Lowe syndrome.
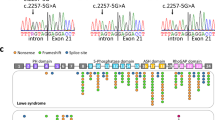
The evolving view of genetic disease transmission should force us to consider that the phenotypic effects also in the so-called monogenic diseases can be the result of the combined action of more than one gene. Taking this view, a possible digenic inheritance was suspected, and we considered OCRL a candidate modifier gene. DNA-sequencing analysis thus revealed an OCRL alteration of the donor splice site consensus sequence – the c.388+3A>G mutation – in our patient and, in the heterozygous state, in his mother (Figure 2a), that has never been described before.
(a) DNA sequencing at the junction between exon 6 and intron 6. The box indicates the nucleotide substitution A>G at the third nucleotide of the donor splice-site consensus sequences (c.388 +3A>G) found in the patient and in his heterozygous mother. (b) cDNA sequencing shows the absence of exon 6 in the patient, but not in his mother. (c) XCI in peripheral leukocytes. Analysis of methylation of HpaII site at the human AR locus. −HpaII, undigested DNA; +HpaII, digested DNA. The same results were obtained using HhaI (data not shown).
The patient’s leukocyte cDNA was amplified by PCR and electrophoresis revealed a fragment 90 bp shorter than expected. On sequencing, this fragment showed an in-frame exon 6 skipping (r.299_388 del exon 6) (Figure 2b). The mutation identified in our patient predictably results in an OCRL1 protein lacking 30 amino acids encoded by exon 6 (p.A100_G129 del), but with the central inositol 5-phosphatase domain and the C-terminal side of the ASH-RhoGAP domain intact. Only the wild-type mRNA was found in the mother’s leukocytes. To ascertain the molecular basis for the monoallelic expression of normal OCRL in the mother, we performed an XCI study on the AR locus. The mother showed a completely skewed X inactivation with a ratio of 100:0 (Figure 2c). DNA analysis of the maternal grandfather revealed no such CLCN5 and OCRL mutations, suggesting that the patient’s X chromosome was inherited from his maternal grandmother, who died of colon cancer at 42 years of age.
The OCRL mutation that we found is located in the first 5′UTR exons, where all frame-shift and splice-site mutations detected in Dent disease 2 patients so far have been clustered. Excepting very few cases, the OCRL mutations associated with Dent disease 2 do not overlap with those causing Lowe syndrome.10, 20, 24 Missense mutations occur in the middle region of the gene (exons 9–15), whereas truncating mutations are found exclusively in the first 7 exons; the OCRL mutations associated with Lowe syndrome are located instead in the 3′ region of the gene (exons 9–22) and involve all three functional OCRL1 domains, whereas frameshift and nonsense mutations cluster in exons 8–23. The different distribution of OCRL mutations in Dent disease 2 and Lowe syndrome suggests a genotype–phenotype correlation that has yet to be thoroughly explored.
Our case is the first to be reported so far of a digenic inheritance with additive effect of Dent’s disease and thus adds to our knowledge of the genetic and phenotypic heterogeneity of Dent’s disease. The phenotype of this patient carrying both OCRL and CLCN5 disease-causing mutations might stem from a positive (synergic) interaction between the two mutations. In fact, he had a severe Dent disease 2 phenotype that was more similar to the syndromic Lowe phenotype.
A likely involvement of epistatic gene–gene interactions between OCRL and CLCN5 in determining a patient’s phenotype was also suggested in a previous case report of ours,24 in which we documented both a silent CLCN5 mutation (P568P) in two brothers, which was predicted, and was demonstrated to strengthen a cryptic splice site, and a missense OCRL mutation (E720D), which was expected to alter the ASH-RhoGAP domain (similar to a typical Lowe mutation). The two brothers’ phenotype was more similar to a Dent disease 2 than to a Lowe syndrome, however, suggesting a negative (antagonistic) interaction between the two mutations.
Our results also indicate that the OCRL1 and ClC-5 proteins cooperate more in cell physiology than was previously believed. Lowe’s group25, 26 showed that OCRL1 is localized to early endosomes and the trans-Golgi apparatus, and is present in clathrin-coated transport intermediates. They also showed that OCRL1 depletion disrupts trafficking at the TGN/endosome interface, suggesting its role in regulating transport between these compartments. The involvement of both ClC-5 and OCRL1 in the assembly of the macromolecular complex implicated in the endocytic pathway might point to the common pathogenic mechanism by means of which renal tubulopathy occurs in Dent’s disease as well as in Lowe syndrome. Our case seems to suggest instead that the cooperation between the two proteins might go beyond renal tubular function. We speculate that, as both the OCRL1 and the ClC-5 proteins are linked through their C-terminal domains to actin dynamics,27, 28, 29 perturbation of the fine mechanisms of cell polarity30, 31 and/or cytokinesis32 might be additional common pathogenic mechanisms.
References
Wrong OM, Nordern AGW, Feest TG : Dent’s disease: a familial renal tubular syndrome with hypercalciuria, tubular proteinuria, rickets, nephrocalcinosis and eventual renal failure. Q J Med 1990; 77: 1086–1087, (Abstract).
Lloyd SE, Pearce SHS, Fisher SE et al: A common molecular basis for three inherited kidney stone diseases. Nature 1996; 379: 445–449.
Thakker RV : Pathogenesis of Dent’s disease and related syndromes of X-linked nephrolithiasis. Kidney Int 2000; 57: 787–793.
Gunther W, Luchow A, Cluzeaud F, Vandewalle A, Jentsch TJ : ClC-5, the chloride channel mutated in Dent’s disease, colocalizes with the proton pump in endocytotically active kidney cells. Proc Natl Acad Sci USA 1998; 95: 8075–8080.
Devuyst O, Christie PT, Courtoy PJ, Beauwens R, Thakker RV : Intra-renal and subcellular distribution of the human chloride channel, CLC-5 reveals a pathophysiological basis for Dent’s disease. Hum Mol Genet 1999; 8: 247–257.
Hara-Chikuma M, Wang Y, Guggino SE, Guggino WB, Verkman AS : Impaired acidification in early endosomes of ClC-5 deficient proximal tubule. Biochem Biophys Res Commun 2005; 329: 941–946.
Picollo A, Pusch M : Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature 2005; 436: 420–423.
Scheel O, Zdebik AA, Lourdel S, Jentsch TJ : Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 2005; 436: 424–427.
Hoopes RR, Shrimpton AE, Knohl SJ et al: Dent Disease with mutations in OCRL1. Am J Hum Genet 2005; 76: 260–267.
Shrimpton AE, Hoopes RR, Knohl SJ et al: OCRL1 mutations in Dent 2 patients suggest a mechanism for phenotypic variability. Nephron Physiol 2009; 112: p27–p36.
Cho HY, Lee BH, Choi HJ, Ha IS, Choi Y, Cheong HI : Renal manifestations of Dent disease and Lowe syndrome. Pediatr Nephrol 2008; 23: 243–249.
Sekine T, Nou K, Iyengar R et al: OCRL1 mutations in patients with Dent disease phenotype in Japan. Pediatr Nephrol 2007; 22: 975–980.
Utsch B, Bökenkamp A, Benz MR et al: Novel OCRL1 mutations in patients with the phenotype of Dent disease. Am J Kidney Dis 2006; 48: e1–e14.
Sethi SK, Ludwig M, Kabra M, Hari P, Bagga A : Vitamin A responsive night blindness in Dent's disease. Pediatr Nephrol 2009; 24: 1765–1770.
Bogdanović R, Draaken M, Toromanović A, Dordević M, Stajić N, Ludwig M : A novel CLCN5 mutation in a boy with Bartter-like syndrome and partial growth hormone deficiency. Pediatr Nephrol 2010; 25: 2363–2368.
Besbas N, Ozaltin F, Jeck N, Seyberth H, Ludwig M : CLCN5 mutation (R347X) associated with hypokalaemic metabolic alkalosis in a Turkish child: an unusual presentation of Dent's disease. Nephrol Dial Transplant 2005; 20: 1476–1479.
Sheffer-Babila S, Chandra M, Speiser PW : Growth hormone improves growth rate and preserves renal function in Dent disease. J Pediatr Endocrinol Metab 2008; 21: 279–286.
Copelovitch L, Nash MA, Kaplan BS : Hypothesis: Dent disease is an underrecognized cause of focal glomerulosclerosis. Clin J Am Soc Nephrol 2007; 2: 914–918.
Frishberg Y, Dinour D, Belostotsky R et al: Dent’s disease manifesting as focal glomerulosclerosis: Is it the tip of the iceberg? Pediatr Nephrol 2009; 24: 2369–2373.
Hichri H, Rendu J, Monnier N et al: From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat 2011; 32: 379–388.
Tosetto E, Ghiggeri GM, Emma F et al: Phenotypic and genetic heterogeneity in Dent’s disease-the results of an Italian collaborative study. Nephrol Dial Transplant 2006; 21: 2452–2463.
Addis M, Loi M, Lepiani C, Cau M, Melis MA : OCRL mutation analysis in Italian patients with Lowe syndrome. Hum Mutat 2004; 23: 524–525.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW : Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239.
Tosetto E, Addis M, Caridi G et al: Locus heterogeneity of Dent’s disease: OCRL1 and TMEM27 genes in patients with no CLCN5 mutations. Pediatr Nephrol 2009; 24: 1967–1973.
Ghanekar Y, Lowe M : Signalling for secretion. Nat Cell Biol 2005; 7: 851–853.
Choudhury R, Diao A, Zhang F et al: Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell 2005; 16: 3467–3479.
Hryciw DH, Wang Y, Devuyst O, Pollock CA, Poronnik P, Guggino WB : Cofilin interacts with ClC-5 and regulates albumin uptake in proximal tubule cell lines. J Biol Chem 2003; 278: 40169–40176.
Kuhbacher A, Dambournet D, Echard A, Cossart P, Pizarro-Cerda J : The phosphatidylinositol 5-phosphatase oculocerebrorenal syndrome of Lowe protein(OCRL) controls actin dynamics during early steps of Listeria monocytogenes infection. J Biol Chem 2012; 287: 13128–13136.
Lowe M : Structure and function of the Lowe syndrome protein OCRL1. Traffic 2005; 6: 711–719.
Moulin P, Igarashi T, Van der Smissen P et al: Altered polarity and expression of H+ATPase without ultrastructural changes in kidneys of Dent's disease patients. Kidney Int 2003; 63: 1285–1295.
Grieve AG, Daniels RD, Sanchez-Heras E et al: Lowe Syndrome protein OCRL1 supports maturation of polarized epithelial cells. PLoS One 2011; 6: e24044.
Dambournet D, Machicoane M, Chesneau L et al: Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat Cell Biol 2011; 13: 981–988.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Addis, M., Meloni, C., Tosetto, E. et al. An atypical Dent’s disease phenotype caused by co-inheritance of mutations at CLCN5 and OCRL genes. Eur J Hum Genet 21, 687–690 (2013). https://doi.org/10.1038/ejhg.2012.225
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.225
Keywords
This article is cited by
-
Prenatal diagnosis of dent disease type I with a nonsense pathogenic variant in CLCN5: a case study
BMC Medical Genomics (2024)
-
A case of Dent disease type 2 with large deletion of OCRL diagnosed after close examination of a school urinary test
CEN Case Reports (2022)
-
Dent disease: classification, heterogeneity and diagnosis
World Journal of Pediatrics (2021)
-
Genetics and phenotypic heterogeneity of Dent disease: the dark side of the moon
Human Genetics (2021)
-
Identification and functional characterization of a hemizygous novel intronic variant in OCRL gene causes Lowe syndrome
Clinical and Experimental Nephrology (2020)