Abstract
Bardet–Biedl syndrome (BBS; OMIM no. 209 900) and Alström syndrome (ALMS; OMIM no. 203 800) are rare, multisystem genetic disorders showing both a highly variable phenotype and considerable phenotypic overlap; they are included in the emerging group of diseases called ciliopathies. The genetic heterogeneity of BBS with 14 causal genes described to date, serves to further complicate mutational analysis. The development of the BBS–ALMS array which detects known mutations in these genes has allowed us to detect at least one mutation in 40.5% of BBS families and in 26.7% of ALMS families validating this as an efficient and cost-effective first pass screening modality. Furthermore, using this method, we found two BBS families segregating three BBS alleles further supporting oligogenicity or modifier roles for additional mutations. We did not observe more than two mutations in any ALMS family.
Similar content being viewed by others
Introduction
Bardet–Biedl syndrome (BBS; OMIM no. 209 900) and Alström syndrome (ALMS; OMIM no. 203 800) are rare, multisystem genetic disorders showing both a highly variable phenotype and considerable phenotypic overlap. BBS, the more prevalent of the two, is genetically heterogeneous and characterised by rod–cone dystrophy, obesity, postaxial polydactyly, cognitive impairment, hypogonadism and renal abnormalities.1 A variety of secondary features have also been reported in BBS patients, including developmental delay, speech disorders, dental anomalies, diabetes mellitus and behavioural problems.
ALMS is a recessively inherited disorder with a complex and variable presentation, including progressive cone–rod dystrophy beginning at birth and leading to blindness, sensorineural hearing loss, obesity and type 2 diabetes in childhood. Most patients develop cardiomyopathy in infancy or adolescence. Pulmonary, hepatic and renal/urological dysfunctions are frequently observed, and systemic fibrosis develops by the end stage of the disease.2
In both BBS and ALMS, the combination of the late onset of some features, such as renal disease, and the existence of other genetic syndromes with similar cardinal manifestations often leads to confusion amongst clinicians and possible misdiagnosis.
To date, one ALMS and 12 BBS genes have been identified.3, 4, 5 In addition, mutations in BBS patients have also been identified in MKS1 and CEP290, genes known to cause other ciliopathic phenotypes such as Meckel syndrome and nephronophthisis.6 Owing to the large number of BBS genes, direct sequencing of all coding exons of these (135 exons for BBS1-12 and 23 exons for ALMS1) is not practical or cost-effective. For this reason, in collaboration with Asper Biotech (http://www.asperbio.com), we developed a BBS–ALMS1 mutation array that can be used to screen a patient sample for known mutations in 10 BBS genes and ALMS1. In this study, we report the analysis of 340 patient samples using the BBS–ALMS1 array confirming its value as a first-pass diagnostic screening test.
Materials and methods
Subjects
DNA samples from 340 patients (205 BBS and 135 ALMS) have been analysed using the BBS–ALMS1 mutation array. Genomic DNA was prepared from blood lymphocytes by standard procedures. The DNA samples were derived from at least 24 different countries. All patients had either a confirmed or suspected diagnosis of BBS or ALMS. The research adhered to the declaration of Helsinki and was approved by the Institutional Review Boards of all participating institutions.
BBS–ALMS1 mutation array
The BBS–ALMS1 mutation array uses arrayed primer extension technology first described in 1996 (Shumaker et al7). This methodology allows the detection of single base substitutions, deletions and insertions. Basically, 5′-modified sequence-specific oligonucleotides are arrayed on a glass slide. In general, these oligonucleotides are designed with their 3′ end immediately adjacent to the variable site. PCR-amplified and fragmented target nucleic acids are annealed to oligonucleotides on the slide, followed by sequence-specific extension of the 3′ ends of primers with dye-labelled nucleotides analogues by DNA polymerase (Supplementary data (so2), http://www.asperbio.com). The latest version of the array contains 253 sequence variants from BBS1-7, BBS9, BBS10, BBS12 and ALMS1 (Supplementary Table 1). In addition to published mutations, intragenic SNPs have also been included on the array. Data from these SNPs could potentially be used to demonstrate linkage to a particular locus and allow a more structured approach to identification of novel BBS and ALMS1 mutations in these samples. Despite this facility, we have not yet formally evaluated the benefits of SNP-based haplotyping in our cohort.
Results
In designing the customised array, we performed chip validation assays obtaining an assay call rate of 99.5%, sensitivity of 99.4% and a specificity of 100%, therefore we assumed the design to be of high quality.
Of the 340 samples that were screened using the mutation array, at least one likely causative mutation was identified in 35% (119/340) of families (Figure 1). In the majority of cases (53 BBS+16 ALMS families) homozygous or compound heterozygous mutations were discovered, therefore, requiring no further analysis (Tables 1 and 2). A summary of polymorphisms that were detected is shown in Supplementary Table 2.
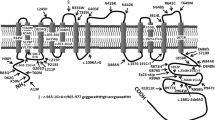
The BBS1 and BBS10 genes are the most common contributors to BBS, being involved in 84.1% (69/82) of the total BBS families with a BBS mutation detected. As it has been reported before, the most frequent disease alleles in north European origin BBS families were p.M390R, identified in all BBS1 families, and p.C91fsX95 (Zughloul and Katsanis)5 in 50% (10/20) of BBS10 families (Table 1). Although the most frequently reported mutation in ALMS1 is c.10775delC (p.T3592fs),8 the most frequently observed ALMS1 mutation in this series was c.11316_11319delAGAG (p.R3772fs), identified in three families.
In two BBS cases (excluding two families with the probable polymorphism p.A242S, BBS6), three bona fide mutations were identified (Table 1). It is, therefore, possible that these are examples of oligogenic inheritance which has previously been reported in BBS.9 No evidence of digenic triallelism was seen in patients with ALMS.
One patient who fulfilled diagnostic criteria for BBS was found to be compound heterozygous for a frame-shift and nonsense mutation in ALMS1. Four patients diagnosed as ALMS, harboured homozygous BBS gene mutations, and one had a heterozygous mutation in BBS10.
Discussion
The BBS–ALMS1 array detected more mutations in the BBS families (40.5%; 83/205) than in ALMS (26.7%; 36/135) families. This finding could be explained by the existence of two ‘common’ mutations in BBS1 and BBS10, identified in 71.1% (59/83) of BBS families; these mutations have been proposed previously for first pass screening.10 In addition, most laboratories testing ALMS families, offer direct ALMS1 sequencing of only exons 16, 10, and part of 8, which would identify the mutations most frequently found in ALMS1. However, other alternative approaches based on massive sequencing are not, at this time, as practical or cost-effective as the genotyping chip.
In our dataset, the array allowed us to identify two mutations in 25.8% (53/205) BBS and 11.9% (16/135) ALMS families. In the cases in whom a single heterozygous BBS or ALMS1 mutation was found, the entire open reading frame of the gene containing that mutation should be screened in order to detect an additional novel causative mutation within that gene. If no mutation is found using the array, the negative results could be explained in a number of ways: (1) the patient has novel mutations in genes that are not represented on the array (BBS8, BBS11, MKS1 or CEP290), (2) the specific mutation the patient carries is not included on the array, or (3) the patient has a mutation in a gene other than BBS or ALMS. A limitation of the array is that, in its present form it does not provide complete coverage of all genes in which mutations have been found in BBS and ALMS patients, nor all of the published mutations in BBS and ALMS. As positions are added on a regular basis, mutations could be picked up if the patient is re-tested at a later date, improving the success rate.
Although the mutations that are on the chip have been described in different populations, the mutation detection rate was higher in European than non-European patients (BBS: 47 vs 3%; ALMS: 19 vs 5%).
This combined syndrome array has the potential to resolve some diagnostic confusion. For example, in this dataset, misdiagnoses in that at least four patients with a clinical diagnosis of ALMS in fact harboured BBS mutations. Conversely, one patient diagnosed phenotypically with BBS had ALMS1 compound heterozygous mutations, c.10775delC (p.T3592fsX3597) and c.11449C>T (p.E3817X), and no BBS mutations.
There are only a few published examples of the presence of a third mutation in a second BBS gene that is necessary for the phenotypic manifestation of BBS,11 possibly owing to the paucity of families with multiple siblings in which the triallelic inheritance can be proven. More often reported however, is the presence of a third heterozygous mutation (at a second BBS locus) behaving as a modifier allele.9 In this study, we were able to ascertain that two probands carried three bone fide BBS mutations, whereas the third heterozygous allele was a nonsense mutation (p.R275X in BBS2; p.Q355X in BBS9). Although there were no siblings in whom either necessity of the third allele could be tested or severity scored, it is likely that the nonsense allele would have a significant effect on the function of the protein complexes that have been described. To date, we have not seen any examples of this in ALMS.
In conclusion, we confirm the use of this array as a helpful and cost-effective first pass screening test for cases of BBS or ALMS and demonstrate it benefit in resolving phenocopy issues between these syndromes. However, the analysis by several approaches of the two more frequent mutations in BBS families (p.M390R, BBS1; p.C91fsX95, BBS10) could be an efficient preliminary step as they were found in 75.6% (60/82) of the patients in which at least one mutation was detected. However, one must be cautious as the most frequent BBS10 mutation in our subset of BBS Spanish patients was p.Y559fsX576, supporting the value of a wide-spectrum approach, such as a genotyping array.
Nevertheless, the development of next generation sequencing technologies will eventually be used for the identification of disease-causing mutation in this group of patients; however, at this time, these approaches are not as practical or cost-effective as the genotyping array.
References
Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA : New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 1999; 36: 437–446.
Marshall JD, Bronson RT, Collin GB et al: New Alström syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med 2005; 165: 675–683.
Hearn T, Renforth GL, Spalluto C et al: Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alström syndrome. Nat Gene 2002; 31: 79–83.
Collin GB, Marshall JD, Ikeda A et al: Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Gene 2002; 31: 74–78.
Zaghloul NA, Katsanis N : Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest 2009; 119: 428–437.
Leitch CC, Zaghloul NA, Davis EE et al: Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet 2008; 40: 443–448.
Shumaker JM, Metspalu A, Caskey CT : Mutation detection by solid phase primer extension. Hum Mutat 1996; 7: 346–354.
Marshall JD, Hinman EG, Collin GB et al: Spectrum of ALMS1 variants evaluation of genotype-phenotype correlations in Alström syndrome. Hum Mutat 2007; 28: 1114–1123.
Badano JL, Kim JC, Hoskins BE et al: Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum Mol Genet 2003; 12: 1651–1659.
Muller J, Stoetzel C, Vincent MC et al: Identification of 28 novel mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet 2010; 127: 583–593.
Katsanis N, Ansley SJ, Badano JL et al: Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 2001; 293: 2256–2259.
Acknowledgements
We thank the BBS and ALMS patients reported here for their willing and continued co-operation in this study. We are grateful to the staff of Asper Biotech, particularly Eneli Oitmaa and Tiina Kantsik for help in the validation stages of the array. We thank Alström Syndrome Canada, Alström Syndrome International, NIH-HD036878 (JDM, GBC, JKN), EsRetNet, Fondo de Investigación Sanitaria (PI06/0049; DV) and Wellcome Trust (PLB) for research support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Pereiro, I., Hoskins, B., Marshall, J. et al. Arrayed primer extension technology simplifies mutation detection in Bardet–Biedl and Alström syndrome. Eur J Hum Genet 19, 485–488 (2011). https://doi.org/10.1038/ejhg.2010.207
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.207
Keywords
This article is cited by
-
Allelic overload and its clinical modifier effect in Bardet-Biedl syndrome
npj Genomic Medicine (2022)
-
A very early diagnosis of Alstrӧm syndrome by next generation sequencing
BMC Medical Genetics (2020)
-
ALMS1 and Alström syndrome: a recessive form of metabolic, neurosensory and cardiac deficits
Journal of Molecular Medicine (2019)
-
Combining targeted panel-based resequencing and copy-number variation analysis for the diagnosis of inherited syndromic retinopathies and associated ciliopathies
Scientific Reports (2018)
-
The phenotypic and molecular genetic spectrum of Alström syndrome in 44 Turkish kindreds and a literature review of Alström syndrome in Turkey
Journal of Human Genetics (2015)