Abstract
NADH:ubiquinone oxidoreductase (complex I; EC 1.6.5.3), the largest respiratory chain complex is composed of 45 proteins and is located at the mitochondrial inner membrane. Defects in complex I are associated with energy generation disorders, of which the most severe is congenital lactic acidosis. We report on four infants from two unrelated families of Jewish Caucasus origin with fatal neonatal lactic acidemia due to isolated complex I deficiency. Whole genome homozygosity mapping, identified a 2.6 Mb region of identical haplotype in the affected babies. Sequence analysis of the nuclear gene encoding for the NDUFS6 mitochondrial complex I subunit located within this region identified the c.344G>A homozygous mutation resulting in substitution of a highly evolutionary conserved cysteine residue by tyrosine. This is the second report of NDUFS6 mutation in humans. Both reports describe three diverse homozygous mutations with variable consequential NDUFS6 protein defects that result in similar phenotype. Our study further emphasizes that NDUFS6 sequence should be analyzed in patients presenting with lethal neonatal lactic acidemia due to isolated complex I deficiency.
Similar content being viewed by others
Introduction
Isolated NADH:ubiquinone oxidoreductase (complex I; EC 1.6.5.3) deficiency is the most common respiratory chain defect, diagnosed in one-third of the patients with OXPHOS dysfunction.1 Five clinical phenotypes are frequently seen in these patients: severe neonatal lactic acidosis, Leigh disease, cardiomyopathy–encephalopathy, hepatopathy–tubulopathy and leukodystrophy with macrocephaly.2, 3
Complex I is the largest and most intricate of the five mitochondrial respiratory chain complexes. It consists of 45 proteins, of which 7 are encoded by the mitochondrial DNA and the remaining by nuclear DNA. Mutations in mitochondrial DNA-encoded genes are detected in only 20% of the children with isolated complex I deficiency4, 5 suggesting that the vast majority of patients harbor mutations in nuclear genes, which encode structural subunits or assembly factors. Till now, extensive sequence determination of candidate genes revealed mutations in 11 structural subunit genes6, 7 and in 4 assembly factors of complex I.8, 9, 10, 11 Still, molecular diagnosis is lacking for more than 50% of the patients12 though the recent description of the mammalian mitochondrial proteome carries great diagnostic promises for many patients.11 The present report is the result of our effort to identify the diseased genes in patients with isolated complex I deficiency originating from consanguineous families using homozygosity mapping.
Case reports
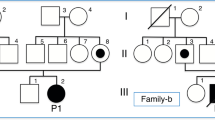
Patient I-1 was born at term following an uneventful pregnancy. Her parents were nonconsanguineous of a Jewish Caucasus descent. On the second day of life she developed encephalopathy and severe lactic acidosis, and expired at the age of 8 days. Thereafter two healthy boys were born. The fourth child, (patient I-2), a male, was born at term, with birth weight of 3460 g (50th centile) and head circumference of 35 cm (50th centile). At 32 h of age he became lethargic, drowsy and refused to eat. Laboratory investigation disclosed normal serum glucose and electrolytes, metabolic acidosis with a pH of 7.1 and serum bicarbonate level of 8 meq/l. Lactate values were elevated both in serum and in CSF, 6.0 mmol/l raising to 11.2 mmol/l within few hours and 8.1 mmol/l, respectively (normal values <2.0 mmol/l). Urine, blood and CSF cultures were sterile. Massive excretion of lactate and Krebs cycle metabolites were noted in the urinary organic acid profile. The severe metabolic acidosis resulted from both elevated serum lactate and the accompanying ketosis. Brain ultrasound and echocardiography were normal. Over the next couple of days, and despite active treatment, the acidosis worsened and the patient became comatose and apneic. His liver was not enlarged, and liver transaminases levels were normal; however, creatine kinase level was 438 IU/l (N<150). Muscle biopsy was performed several hours before death and the activities of the five enzymatic complexes of the mitochondrial respiratory chain in muscle mitochondria were determined.
Patient II-1 was the first daughter of a Jewish Caucasus nonconsanguineous couple. She was born after an uneventful pregnancy and delivery, with normal Apgar score and birth weight of 2400 g. At the age of 6 days she presented with apathy, tachypnea and severe metabolic acidosis and died within several hours. Blood, urine and CSF cultures were sterile.
Her sister, patient II-2, was a full term seemingly healthy newborn, with birth weight of 2330 g. At the age of 6 days she presented with symptoms resembling those of her deceased sister. On admission she was pale, tachypneic, severely hypotonic and unresponsive to any stimuli. There were no dysmorphic features and her liver and spleen were not enlarged. Laboratory studies revealed severe metabolic acidosis with blood pH of 7.02 and serum bicarbonate of 8.6 mmol/l. Plasma lactate was 16.8 mmol/l, and the CSF lactate level was 18.4 mmol/l (normal values <2.0 mmol/l). Serum electrolytes as well as liver transaminases and creatine kinase levels were normal. Serum ammonia was elevated ranging between 78 and 275 μmol/l (normal values 5–45 μmol/l). The patient died at the age of 8 days. Muscle and liver biopsies were performed immediately after death. Light microscopy of the muscle was unremarkable, whereas electron microscopy showed several cells with microvesicular steatosis and several atypical mitochondria with distorted cristae. Liver biopsy revealed nonspecific micro- and macrosteatosis.
Methods
The enzymatic activities of the mitochondrial respiratory-chain complexes in mitochondria isolated from muscle were determined as previously described.13
To localize the mutated gene, homozygosity mapping was performed using GeneChip Human Mapping 250K Nsp Array of Affymetrix. The analysis of the 250 000 SNPs, was performed as previously described.14 DNA samples of the only available patients, I-2 and II-2, were analyzed, and homozygous regions >4.5 Mb were sought, as smaller homozygous regions were reported to occur at random, even in the offspring of nonconsanguineous couples.15 The study was approved by the Hadassah Medical Center Ethical Review Committee.
Results
Enzymatic activities of the five respiratory chain complexes were determined in isolated muscle mitochondria from patients I-2 and II-2, by standard spectrophotometric methods and normalized to citrate synthase.13 The activity of complex I in muscle mitochondria, determined as rotenone sensitive NADH:cytochrome c oxidoreductase (complex I+III), NADH:ubiquinone reductase and NADH:ferricyanide reductase, was reduced to 22, 42 and 45% of the control mean, respectively, in patient I-2, and to 7, 38 and 56%, respectively, in patient II-2. The activities of the other four respiratory chain complexes were within the normal range.
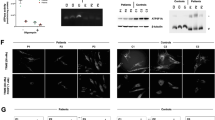
The analysis of the 250 000 SNPs disclosed homozygosity at two regions >4.5 Mb on chromosome 10 (51.46–59.18 Mb) and on chromosome 12 (96.40–109.77 Mb) in patient II-2 sample. There were no complex I genes within these genomic regions and the only mitochondrial annotated gene which we considered relevant, MTERFD3, was sequenced and was found normal. Patient I-2 was homozygous for four >4.5 Mb regions (chromosome 5: 126.11–134.64, chromosome 11: 8.05–15.77, chromosome 12: 118.19–124.47 Mb, chromosome19: 0–6.63 Mb), of which chromosome 5 region could reasonably be excluded because his healthy brother had the same polymorphic micro satellite markers haplotype. The sequences of the two known complex I genes within these regions, NDUFS7 and NDUFA11 genes, were normal. As the two families originated from the same ethnic group and although reportedly unrelated, we assumed a common founder mutation and have compared the 250 000 SNP genotypes of the two patients. This has resulted in the identification of a 2.6 Mb region on chromosome 5 at nucleotide 81 949–2 602 852 (markers rs7704488–rs6885850) where the two patients shared an identical haplotype over 182 SNPs. Within this region there were 39 ORFs, including the NDUFS6 gene which encodes the 13 kDa subunit of complex I. Sequence analysis of the four exons and their flanking intronic sequences disclosed the transition c.344G>A mutation in a homozygous state in both patients, which results in substitution at codon 115 of a highly conserved cysteine by tyrosine (Figure 1). The mutation is predicted deleterious in silico (SIFT – sorting intolerant from tolerant, http://blocks.fhcrc.org/sift/SIFT.html).
(a) Sequence analysis of NDUFS6 gene in our patients reveals a G to A substitution at nucleotide position 344. Arrow indicates the mutation. (b) The region of the NDUFS6 peptide containing the C115Y mutation is aligned with sequences of NDUFS6 proteins from other species showing the highly conserved residue.
Finally we have determined the prevalence of the mutation among Caucasus Jews. As the mutation abolished a cleavage site for the restriction enzyme HpyCH4III, we used this enzyme for carrier detection. Among the 48 nonrelated individuals of Jewish Caucasus origin we identified two carriers.
Discussion
Isolated complex I deficiency manifests clinically as Leigh disease, cardiomyopathy–encephalopathy, hepatopathy–tubulopathy, leukodystrophy with macrocephaly and severe neonatal lactic acidosis.2, 3 Leigh and Leigh-like syndromes are the commonest presentations of complex I deficiency in young age, and mutations in six nuclear genes (NDUFS1, NDUFS3, NDUFS4, NDUFS7, NDUFS8 and NDUFV1) have been identified in these patients.6 Early-onset encephalo–cardiomyopathy has been previously reported in patients harboring mutations in complex I genes NDUFS2 and NDUFV2.16, 17 Till now, the only molecular cause for fatal infantile lactic acidemia among complex I-deficient patients was a mutation in the NDUFA11 gene.7
Complex I has an L-shaped structure with a hydrophobic arm embedded in the lipid membrane and a hydrophilic, peripheral arm protruding into the matrix.18 NDUFS6 encodes a 13 kDa subunit of complex I, which is part of the peripheral arm and is localized in the iron–sulfur fraction of the complex. Mutations in the NDUFS6 gene were previously identified in only two patients.19 Similar to our patients, they presented with fatal infantile lactic acidemia, suggesting that analysis of NDUFS6 sequence should be considered in patients presenting with lethal neonatal lactic acidemia and isolated complex I deficiency.
The accumulating data indicate that NDUFS6 is important but not essential for complex I assembly. Although the mutations described previously were deleterious (splice site mutation resulting in a frame shift and a large genomic deletion), fully assembled 900 kDa complex I was formed along with a smaller ∼750 kDa subassembly intermediate.19 This complex I subassembly was shown to be associated with complex III in a supercomplex with a smaller molecular weight compared with that found in normal mitochondria.20 Normally assembled complex I was generated on reintroduction of NDUFS6 into mitochondria of NDUFS6-deficient patients suggesting that NDUFS6 is involved in the final stage of the modular process of complex I assembly.20 As normal-sized complex I assembly is formed even in the complete absence of an NDUFS6 subunit, we speculate that the severe clinical phenotype reported in all NDUFS6-mutated patients is the result of an impaired interaction of complex I with complex III. This is supported by the residual activity of NADH:cytochrome c oxidoreductase (I+III) which was the most severely affected in our patients' muscles.
The Caucasus region, formerly belonged to the Soviet Union, was divided in the post-Soviet era into several independent countries and autonomic republics. The Jewish community in the Caucasus region is believed to originate from today's southern Iran in 722 BC. This community was genetically isolated for more than 2500 years by its language and religious practice. Most of the Caucasus Jews immigrated to Israel during 1970–1990. The relatively small size of the common homozygous genomic region and the fact that both couples firmly insisted on the lack of known consanguinity attests to the ancient nature of the causative founder mutation. Although the number of anonymous controls is small, our results suggest that the NDUFS6 mutation c.344G>A is of considerable prevalence in this ethnic group. Based on our findings we suggest that this mutation be screened in Jewish couples originating from this region. This would enable a better estimation of the carrier rate and identify couples at risk.
This report also illustrates the power of homozygosity mapping in the molecular diagnosis of patients with mitochondrial respiratory chain defects, even when only a single patient in a family is available for analysis. Our strategy of routine SNP-chip analysis of all the mitochondrial patients whose parents originate from the same ethnic origin proved useful. Notably, the patients reported herein, I-2 and II-2, were born and analyzed four years apart. It was only the meticulous comparison of the newly analyzed patient to all former samples, which allowed the detection of the NDUFS6 mutation.
Finally, we conclude that genomic regions as short as 2.5 Mb should not be ignored when the parents are of the same ethnicity and an ancient common ancestor is assumed, even if they are reportedly unrelated.
References
Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR : Respiratory chain complex I deficiency: an under diagnosed energy generation disorder. Neurology 1999; 52: 1255–1264.
Pitkanen S, Feigenbaum A, Laframboise R, Robinson BH : NADH-coenzyme Q reductase (complex I) deficiency: heterogeneity in phenotype and biochemical findings. J Inherit Metab Dis 1996; 19: 675–686.
Loeffen JL, Smeitink JA, Trijbels JM et al: Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat 2000; 15: 123–134.
Lebon S, Chol M, Benit P et al: Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J Med Genet 2003; 40: 896–899.
McFarland R, Kirby DM, Fowler KJ et al: De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol 2004; 55: 58–64.
Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA : Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis 2006; 29: 499–515.
Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O : Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol 2008; 63: 405–408.
Ogilvie I, Kennaway NG, Shoubridge EA : A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest 2005; 115: 2784–2792.
Dunning CJ, McKenzie M, Sugiana C et al: Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 2007; 26: 3227–3237.
Saada A, Edvardson S, Rapoport M et al: C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet 2008; 82: 32–38.
Pagliarini DJ, Calvo SE, Chang B et al: A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008; 134: 112–123.
Thorburn DR, Sugiana C, Salemi R et al: Biochemical and molecular diagnosis of mitochondrial respiratory chain disorders. Biochim Biophys Acta 2004; 1659: 121–128.
Saada A, Bar-Meir M, Belaiche C, Miller C, Elpeleg O : Evaluation of enzymatic assays and compounds affecting ATP production in mitochondrial respiratory chain complex I deficiency. Ann Biochem 2004; 335: 66–72.
Edvardson S, Shaag A, Kolesnikova O et al: Deleterious mutation in the mitochondrial arginyl-tRNA synthetase gene is associated with ponto-cerebellar hypoplasia. Am J Hum Genet 2007; 81: 857–862.
Woods CG, Cox J, Springell K et al: Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet 2006; 78: 889–896.
Loeffen J, Elpeleg O, Smeitink JA et al: Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol 2001; 49: 195–201.
Benit P, Beugnot R, Chretien D et al: Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum Mutat 2003; 21: 582–586.
Grigorieff N : Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol 1998; 277: 1033–1046.
Kirby DM, Salemi R, Sugiana C et al: NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest 2004; 114: 837–845.
Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT : Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol 2007; 27: 4228–4237.
Acknowledgements
This work was funded by a research grant from the Israeli Ministry of Health (to OE and AS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Spiegel, R., Shaag, A., Mandel, H. et al. Mutated NDUFS6 is the cause of fatal neonatal lactic acidemia in Caucasus Jews. Eur J Hum Genet 17, 1200–1203 (2009). https://doi.org/10.1038/ejhg.2009.24
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.24
Keywords
This article is cited by
-
NDUFS6 related Leigh syndrome: a case report and review of the literature
Journal of Human Genetics (2019)
-
New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre
Journal of Translational Medicine (2016)
-
Natural disease course and genotype‐phenotype correlations in Complex I deficiency caused by nuclear gene defects: what we learned from 130 cases
Journal of Inherited Metabolic Disease (2012)