Abstract
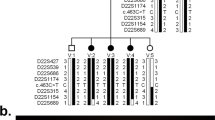
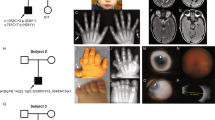
A number of different eye disorders with the presence of early-onset glaucoma as a component of the phenotype have been mapped to human chromosome 6p25. These disorders have been postulated to be either allelic to each other or associated with a cluster of tightly linked genes. We have identified two primary congenital glaucoma (PCG) patients with chromosomal anomalies involving 6p25. In order to identify a gene involved in PCG, the chromosomal breakpoints in a patient with a balanced translocation between 6p25 and 13q22 were cloned. Cloning of the 6p25 breakpoint led to the identification of two candidate genes based on proximity to the breakpoint. One of these, FKHL7, encoding a forkhead transcription factor, is in close proximity to the breakpoint in the balanced translocation patient and is deleted in a second PCG patient with partial 6p monosomy. Furthermore, FKHL7 was found to harbour mutations in patients diagnosed with Rieger anomaly (RA), Axenfeld anomaly (AA) and iris hypoplasia (IH). This study demonstrates that mutations in FKHL7 cause a spectrum of glaucoma phenotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Quigley, H.A. Number of people with glaucoma worldwide. Br. J. Ophthalmol. 80, 389–393 (1996)
Leske, M.C. The epidemiology of open-angle glaucoma: a review. Am. J. Epidemiol. 118, 166–191 (1983)
Sommer, A. et al. Relationship between intraocular pressure and primary open angle glaucoma among white and black Americans. The Baltimore Eye Survey. Arch. Ophthalmol. 109, 1090–1095 (1991)
Stone, E.M. et al. Identification of a gene that causes primary open angle glaucoma. Science 275, 668–670 ( 1997)
Stoilov, I., Akarsu, A.N. & Sarfarazi, M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum. Mol. Genet. 6, 641–647 (1997)
Semina, E.V. et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nature Genet. 14, 392–399 (1996)
Alward, W.L.M. et al. Autosomal Dominant Iris Hypoplasia Is Caused By a Mutation In the Rieger-Syndrome (Rieg/Pitx2) Gene. Am. J. Ophthalmol. 125, 98–100 (1998)
Akarsu, A.N. et al. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum. Mol. Genet. 5, 1199–1203 (1996)
Phillips, J.C. et al. A second locus for Rieger syndrome maps to chromosome 13q14. Am. J. Hum. Genet. 59, 613–619 (1996)
Stoilova, D. et al. Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics 36, 142 –150 (1996)
Wirtz, M.K. et al. Mapping a gene for adult-onset primary en-angle glaucoma to chromosome 3q . Am. J. Hum. Genet. 60, 296–304 (1997)
Mears, A.J., Mirzayans, F., Gould, D.B., Pearce, W.G. & Walter, M.A. Autosomal dominant iridogoniodysgenesis anomaly maps to 6p25. Am. J. Hum. Genet. 59, 1321–1327 (1996)
Gould, D.B., Mears, A.J., Pearce, W.G. & Walter, M.A. Autosomal dominant Axenfeld-Rieger anomaly maps to 6p25. Am. J. Hum. Genet. 61, 765–768 (1997)
Jordan, T., Ebenezer, N., Manners, R., McGill, J. & Bhattacharya, S. Familial glaucoma iridogoniodysplasia maps to a 6p25 region implicated in primary congenital glaucoma and iridogoniodysgenesis anomaly. Am. J. Hum. Genet. 61, 882–888 (1997)
Graff, C., Jerndal, T. & Wadelius, C. Fine mapping of the gene For autosomal dominant juvenile-onset glaucoma with iridogoniodysgenesis In 6p25-Tel. Hum. Genet. 101, 130–134 (1997)
Alward, W.L.M., Johnson, A.T., Nishimura, D.Y., Sheffield, V.C. & Stone, E.M. Molecular genetics of glaucoma: current status. J. Glaucoma 5, 276–284 (1996)
Murray, J.C. et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC). Science 265, 2049 –2054 (1994)
Dorin, J.R. et al. Gene targeting for somatic cell manipulation: rapid analysis of reduced chromosome hybrids by Alu-PCR fingerprinting and chromosome painting. Hum. Mol. Genet. 1, 53–59 ( 1992)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 ( 1990)
Gish, W. & States, D.J. Identification of protein coding regions by database similarity search. Nature Genet. 3, 266–272 (1993)
Currie, H.L., Lightfoot, J. & Lam, J.S. Prevalence of GCA, a gene involved in synthesis of A-band common antigen polysaccharide in Pseudomonas aeruginosa. Clin. Diagn. Lab. Immunol. 2, 554–562 (1995)
Li, Y. et al. Analysis of 43 kb of the Chlorella virus PBCV-1 330-kb genome: map positions 45 to 88. Virology 212, 134–150 (1995)
Stevenson, G., Andrianopoulos, K., Hobbs, M. & Reeves, P.R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178, 4885–4893 (1996)
Bonin, C.P., Potter, I., Vanzin, G.F. & Reiter, W.D. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94, 2085–2090 (1997)
Pierrou, S., Hellqvist, M., Samuelsson, L., Enerback, S. & Carlsson, P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 13, 5002–5012 (1994)
Larsson, C. et al. Chromosomal localization of six human forkhead genes, freac-1 (FKHL5), -3 (FKHL7), -4 (FKHL8), -5 (FKHL9), -6 (FKHL10), and -8 (FKHL12). Genomics 30, 464–469 ( 1995)
Xu, Y., Mural, R., Shah, M. & Uberbacher, E. Recognizing exons in genomic sequence using GRAIL II. Genet. Engin. 16, 241–253 (1994)
Uberbacher, E.C. & Mural, R.J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc. Natl. Acad. Sci. USA 88, 11261–11265 (1991)
Glaser, T., Walton, D.S. & Maas, R.L. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nature Genet. 2, 232–239 (1992)
Jordan, T. et al. The human PAX6 gene is mutated in two patients with aniridia. Nature Genet. 1, 328–332 (1992)
Kozak, M. Interpreting cDNA sequences: some insights from es on translation. Mamm. Genome 7, 563–574 (1996)
Kozak, M. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8, 197–225 (1992)
Attree, O. et al. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358 , 239–242 (1992)
Fantes, J. et al. Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype. Hum. Mol. Genet. 4, 415–422 (1995)
Milot, E., Fraser, P. & Grosveld, F. Position effects and genetic disease. Trends Genet. 12, 123–126 (1996)
Galili, N. et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma . Nature Genet. 5, 230–235 (1993)
Puck, J.M., Nussbaum, R.L. & Conley, M.E. Carrier detection in X-linked severe combined immunodeficiency based on patterns of X chromosome inactivation. J. Clin. Invest. 79, 1395–1400 ( 1987)
Nussbaum, R.L., Airhart, S.D. & Ledbetter, D.H. Expression of the fragile (X) chromosome in an interspecific somatic cell hybrid. Hum. Genet. 64, 148 –150 (1983)
Bassam, B.J., Caetano-Anolles, G. & Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196, 80–83 (1991)
Hudson, T.J. et al. An STS-based map of the human genome. Science 270, 1945–1954 (1995)
Laitinen, J., Samarut, J. & Holtta, E. A nontoxic and versatile protein salting-out method for isolation of DNA. Biotechniques 17, 316, 318, 316–322 (1994)
Newton, C.R. et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503 –2516 (1989)
Clark, K.L., Halay, E.D., Lai, E. & Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 ( 1993)
Acknowledgements
We are grateful to the patients and their families for their participation in this study. We would also like to thank D. Crouch for family recruitment, G. Beck, S. Brown, R. Hockey, L. Law and N. Meyer for technical assistance, S. Fischer for cosmids in the 13q22 region and J. Lin for providing mouse embryos and adult tissues. This work was supported by a grant from the Knight's Templar Eye Foundation (D.Y.N.) and NIH grant R01-EY-10564 (V.C.S. and E.M.S.). We also thank the Roy J. Carver Charitable Trust and the Glaucoma Research Foundation for support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishimura, D., Swiderski, R., Alward, W. et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19, 140–147 (1998). https://doi.org/10.1038/493
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/493
This article is cited by
-
Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci
Nature Genetics (2023)
-
Axenfeld–Rieger syndrome: orthopedic and orthodontic management in a pediatric patient: a case report
Head & Face Medicine (2022)
-
The role of LncRNA MCM3AP-AS1 in human cancer
Clinical and Translational Oncology (2022)
-
EZH2 regulates expression of FOXC1 by mediating H3K27me3 in breast cancers
Acta Pharmacologica Sinica (2021)
-
CPAMD8 loss-of-function underlies non-dominant congenital glaucoma with variable anterior segment dysgenesis and abnormal extracellular matrix
Human Genetics (2020)