Abstract
Read-through fusion transcripts that result from the splicing of two adjacent genes in the same coding orientation are a recently discovered type of chimeric RNA. We sought to determine if read-through fusion transcripts exist in breast cancer. We performed paired-end RNA-seq of 168 breast samples, including 28 breast cancer cell lines, 42 triple negative breast cancer primary tumors, 42 estrogen receptor positive (ER+) breast cancer primary tumors, and 56 non-malignant breast tissue samples. We analyzed the sequencing data to identify breast cancer associated read-through fusion transcripts. We discovered two recurrent read-through fusion transcripts that were identified in breast cancer cell lines, confirmed across breast cancer primary tumors, and were not detected in normal tissues (SCNN1A-TNFRSF1A and CTSD-IFITM10). Both fusion transcripts use canonical splice sites to join the last splice donor of the 5′ gene to the first splice acceptor of the 3′ gene, creating an in-frame fusion transcript. Western blots indicated that the fusion transcripts are translated into fusion proteins in breast cancer cells. Custom small interfering RNAs targeting the CTSD-IFITM10 fusion junction reduced expression of the fusion transcript and reduced breast cancer cell proliferation. Read-through fusion transcripts between adjacent genes with different biochemical functions represent a new type of recurrent molecular defect in breast cancer that warrant further investigation as potential biomarkers and therapeutic targets. Both breast cancer associated fusion transcripts identified in this study involve membrane proteins (SCNN1A-TNFRSF1A and CTSD-IFITM10), which raises the possibility that they could be breast cancer-specific cell surface markers.
Similar content being viewed by others
Introduction
Fusion genes with oncogenic activity were first identified in hematologic malignancies, where chromosomal translocations frequently join two genes that result in an aberrant protein product [1, 2]. These fused genes have been valuable prognostic markers and therapeutic targets [3]. The therapeutic value of identifying fusion genes is exemplified by the development of selective inhibitors targeted to the ABL kinase involved in the BCR–ABL fusion that is present in 95 % of patients with chronic myelogenous leukemia [1, 2, 4]. Most recurrent fusion genes have been identified in leukemias, lymphomas, and soft tissue sarcomas where cytogenetic approaches to detect chromosomal aberrations using spectral karyotyping, fluorescent in situ hybridization, and flow cytometry have been developed [5]. Cytogenetic approaches to detect fusion genes in the more common forms of cancer, epithelial tumors, are hampered by the poor chromosome morphology, complex karyotypes, and cellular heterogeneity that typify these tumors, although it has been posited that fusion genes are likely drivers of oncogenesis in these tumors as well [3, 5, 6]. Until recently, the most prevalent recurrent fusion genes identified in breast cancer were the ETV6-NTRK3 fusion in secretory breast carcinoma, a rare subtype of infiltrating ductal carcinoma [7] and the MYB-NFIB fusion in adenoid cystic carcinomas, another rare form of breast cancer [8]. Recently, genome-wide microarray profiling, the whole genome sequencing and the whole transcriptome sequencing have made it possible to systematically identify fusion genes in solid tumors. With these methods, recurrent fusions that contribute to malignancy have been identified in prostate cancer (e.g., TMPRSS2 fused to ETS family transcription factors [9–11]), in lung cancer (EML4–ALK [12]), and in breast cancer (MAST kinases fused to NOTCH family genes [13]). New technologies and informatics approaches are enabling the identification of recurrent fusion genes in more common epithelial cancers that may serve as valuable biomarkers and drug targets [13–19].
In addition to fusion genes created by genomic rearrangements, fusion transcripts created by cis- and trans-splicing of mRNA, in the absence of a DNA rearrangements, have been detected by sequencing cDNA clone libraries and performing RNA-seq [20]. These chimeric RNAs have been detected at low levels in expressed sequence tag (EST) libraries [21–23] and low levels across benign and malignant samples [6, 20, 24]. One particularly prevalent class of chimeric RNAs involves adjacent genes in the same coding orientation that are spliced together to form an in-frame chimeric transcript that spans both genes. In the recent literature, these have been referred to as read-through gene fusions, transcription-induced chimeras, co-transcription of adjacent genes coupled with intergenic splicing (CoTIS), or conjoined genes. Several of these read-through fusion transcripts have been identified specifically in prostate cancer and are associated with cellular proliferation and disease progression [25–33]. Recurrent read-through transcripts have not yet been characterized in breast cancer. We used paired-end RNA-seq to identify two novel recurrent read-through fusion transcripts associated with breast cancer, and we used genomic DNA sequencing, qPCR, cDNA clone sequencing, small interfering RNA (siRNA) knockdown, and Western blots to further confirm and characterize these fusion transcripts.
Results
Identification of read-through fusion transcripts in breast cancer cell lines
While recent studies have reported recurrent fusion genes in breast cancer that are the result of genomic rearrangements [13, 15, 16, 18, 34], recurrent read-through fusion transcripts in breast cancer have not been previously characterized. We performed RNA-seq [35] on 28 breast cancer cell lines to identify candidate read-through fusion transcripts. We used the ChimeraScan software package to identify read-through transcripts in the RNA-seq data [36]. There were 6 candidate read-through fusion transcripts that were supported by at least 10 read pairs that connect adjacent genes and at least one sequencing read that spanned the fusion junction in more than two breast cancer cell lines (SIDT2-TAGLN, CTBS-GNG5, CLTC-VMP1, MFGE8-HAPLN3, SCNN1A-TNFRSF1A, CTSD-IFITM10; Table1).
Confirmation of candidate fusion transcripts in primary breast tumors
To determine if the read-through fusion transcripts detected in breast cancer cell lines were present in primary breast tumors, we performed RNA-seq [35] on 42 fresh frozen triple negative breast cancer (TNBC) primary tumors and 42 fresh frozen estrogen receptor positive (ER+) breast cancer primary tumors. We again used the ChimeraScan software package to identify read-through transcripts in the RNA-seq data [36]. Five of the candidate fusion transcripts were detected with at least one fusion junction-spanning read in the primary tumors (SIDT2-TAGLN, CTBS-GNG5, MFGE8-HAPLN3, SCNN1A-TNFRSF1A, CTSD-IFITM10; Table 1).
Tumor specificity of fusion transcripts
To determine if the read-through fusion transcripts were associated with breast cancer, or whether they were present in normal tissues, we then performed RNA-seq [35] on 21 uninvolved breast tissue samples that were adjacent to TNBC tumors, 30 uninvolved breast tissue samples that were near ER+ breast tumors, and five normal breast tissue samples that were collected from cancer-free patients during reduction mammoplasty procedures. We also analyzed RNA-seq data from 13 normal human tissues collected by the Illumina Human Body Map 2.0 project, which includes adipose, brain, breast, colon, heart, kidney, liver, ovary, prostate, skeletal muscle, testes, thyroid, and white blood cells [15]. We again used the ChimeraScan software package to identify read-through transcripts in the RNA-seq data [36]. The SIDT2-TAGLN and CTBS-GNG5 fusion transcripts were detected at a high frequency in a variety of normal tissues (Table 1).
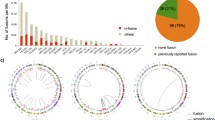
The remaining three fusion transcripts we detected exclusively in breast tumor and normal tissue are MFGE8-HAPLN3, SCNN1A-TNFRSF1A and CTSD-IFITM10. We used Fisher’s Exact test to determine if the read-through fusion transcripts were significantly overrepresented in the breast cancer samples compared to the non-cancer breast samples. We found that SCNN1A-TNFRSF1A and CTSD-IFITM10 were significantly associated with breast cancer (p < 0.05; Table 1). The fusion junction-spanning reads for these read-through fusion transcripts are depicted in Fig. 1, and the number of fusion junction-spanning reads in each sample is reported in Supplemental Table 1. These fusions were present in both ER+ breast cancer and TNBC, and they are frequent events. In our cohorts the breast cancer associated fusion transcripts were detected in 46 % (13/28) of the breast cancer cell lines, 29 % (12/42) of the TNBC primary tumors, and 19 % (8/42) of the ER+ breast cancer primary tumors.
Breast cancer associated read-through fusion transcripts. Two breast cancer associated read-through fusion transcripts, SCNN1A-TNFRSF1A (a) and CTSD-IFITM10 (b), were detected in paired-end RNA-seq performed on breast cancer cell lines and primary tumors and were not detected in a variety of non-neoplastic human tissues. The 5′ gene partner is depicted in green, and the 3′ gene partner is depicted in red. The fusion transcripts use endogenous splice sites to fuse the two transcripts and the angled black lines indicate which exons flank the fusion junction to result in the chimeric transcript. RNA-seq reads that span the fusion junction are depicted above the gene models and the sequence from the 5′ partner is in green text and the sequence from the 3′ partner is in red text. The intergenic chromosomal distance between the fusion partners is denoted in kilobase pairs (kbp). Breast cancer cell line cDNA was PCR amplified using primers in the distal ends of the partner genes, and clones were sequenced. The alignment of the cDNA to the genome and the canonical gene models at this locus are depicted for SCNN1A-TNFRSF1A (c) and CTSD-IFITM10 (d). Both fusion transcripts include all of the canonical exons and splice sites of the partner genes up to the fusion junction and the fusion junction maintains the reading frame of the canonical transcripts
The CTSD-IFITM10 fusion transcript was not detected in any normal tissue RNA-seq data. To determine if the CTSD-IFITM10 fusion is transcribed in normal tissue below the level of detection of RNA-seq, we performed qPCR using primers that flank the fusion junction (Fig. 4a) in 9 normal breast tissue samples including 3 non-malignant tissues samples adjacent to TNBC tumors, 2 non-malignant tissues adjacent to ER+ tumors, and 4 normal breast tissue samples from reduction mammoplasty procedures. The expression of the fusion transcript in normal samples was compared to the expression of the fusion transcript in MDA-MB-468, a cell line in which 9 fusion junction-spanning reads were detected by RNA-seq. The fusion transcript expression measurements in the normal samples were near the limit of detection of our qPCR assay, and were an average of 84 fold lower than the expression in the positive control cell line (Supplemental Fig. 1). These results are consistent with the lack of expression observed in the normal tissue RNA-seq data.
Structure and expression of read-through fusion messages
To determine which exons are included in the breast cancer associated fusion transcripts, we PCR amplified the fusion transcript from breast cancer cell line cDNA using forward primers in the 5′ gene exons and reverse primers in the 3′ gene exons. We then cloned and sanger sequenced the PCR products from the most distal primers to determine the full coding sequence of the fusion transcripts. Both SCNN1A-TNFRSF1A and CTSD-IFITM10 included all canonical exons and splice sites of the partner genes up to the fusion junction, and the coding sequence is in-frame across the fusion junction (Fig. 1).
For the read-through fusion mRNA to be transcribed, RNA polymerase would begin in the promoter of the 5′ gene, continue across the intergenic region and terminate after the 3′ UTR of the 3′ gene. This is possible for these fusion transcripts, because the intergenic region between the genes is small for both loci (4.8 kbp between SCNN1A and TNFRSF1A, and 2.2 kbp between CTSD and IFITM10). Additionally, the genomic distance from the start of the 5′ gene partner to the end of 3′ gene partner is less than the average genomic distance traversed by RNA polymerase II for canonical genes in the human genome (48 kbp for SCNN1A-TNFRSF1A, 31 kbp for CTSD-IFITM10, 56 kbp for average gene length in human genome).
Both fusion transcripts use canonical splice sites to join the last splice donor of the 5′ gene to the first splice acceptor of the 3′ gene. This splicing pattern skips the last exon of the 5′ gene and the first exon of the 3′ gene (Fig. 1). In order for this product to form, the 5′ gene’s terminal exon splice acceptor site has been skipped, which results in the usage of the next available splice acceptor residing in the adjacent 3′ gene. To determine whether a mutation or a deletion at the 5′ gene’s terminal exon is associated with the formation of these read-through fusion transcripts, we sequenced 200 bp of genomic DNA surrounding the skipped splice acceptor site. We did not identify any mutations associated with the presence of the fusion transcripts and we observed both alleles of heterozygous SNPs at expected frequencies. These results indicate that neither fusion transcript is associated with genomic DNA mutations or deletions of the skipped last exon of the 5′ gene.
An alternative hypothesis is that the kinetics of transcription at these loci are skewed to favor inter-gene splicing of the read-through fusion transcript before canonical splicing and 3′ cleavage of the upstream gene occurs. We calculated the fraction of reads near the fusion junction that include sequence from the fusion transcript rather than the un-fused canonical transcripts. This fraction reflects the abundance of the chimeric transcript relative to the canonical isoform (Fig. 2a). Only a small fraction of the transcripts from the 5′ gene include the fusion, and a significantly higher fraction of transcripts from the 3′ gene are fusion transcripts (Mann–Whitney test: SCNN1A vs TNFRSF1A p = 0.0247, and CTSD vs IFITM10 p < 0.0001). This indicates that a larger proportion of the transcription of the 3′ partner is created from read-through transcripts beginning at the 5′ gene promoter. Higher expression of the 5′ gene could lead to run-on transcription into the adjacent 3′ gene. We examined the expression of the 5′ fusion partner gene but found that there was no difference in expression levels between samples with and without the fusion. This indicates that the steady state expression level of the 5′ gene is not associated with the presence of these fusions (Fig. 2b). In summary, these breast cancer associated read-through fusion transcripts, which account for a significant portion of the 3′ gene’s expression, are independent of the 5′ gene’s expression level.
Expression of genes involved in breast cancer associated read-through fusion transcripts. a We computed the fraction of reads that include sequence from the fusion transcript rather than the un-fused canonical transcript. The fraction of fusion transcript reads for 5′ fusion partners are indicated in green, and the 3′ fusion partners are denoted in red for each of the samples. Mean and standard error of the mean are depicted in black. Less than 20 % of the 5′ fusion partners’ transcripts include the fusion sequence, indicating that most of the transcripts from the 5′ gene are not fused. A significantly larger fraction of the 3′ gene’s transcripts contain the fusion sequence (Mann–Whitney test: SCNN1A vs TNFRSF1A p = 0.0247, and CTSD vs IFITM10 p < 0.0001). b There is not a significant difference in the expression levels of the 5′ fusion partner between samples with or without the read-through fusion transcript (labeled fused and not fused, respectively). This indicates that increased expression of the 5′ fusion partner is not sufficient to induce read-through fusion transcripts that include the 3′ gene
Detection of fusion proteins
Both of the breast cancer associated read-through fusion transcripts we identified involved genes that encode membrane proteins. These proteins’ functions rely on their correct placement in the membrane and correct participation in protein complexes. SCNN1A is an alpha subunit of nonvoltage-gated, amiloride-sensitive, sodium channels [37]. It is fused to TNFRSF1A, a tumor necrosis factor-alpha receptor that activates NF-κB, mediates apoptosis, and regulates inflammatory responses [38]. CTSD is a lysosomal aspartyl protease that also functions as a secreted protein that binds membrane receptors and has previously been associated with breast cancer [39]. It is fused to IFITM10, a member of a family of membrane proteins that are induced by interferon and are involved in cell proliferation and cell adhesion [40]. These read-through fusion transcripts join genes that have disparate functions, suggesting that a fused protein could impair normal function or localization in breast cancer.
We predicted the length of the fusion protein based upon the fusion transcript sequence, and used Western blots with an antibody raised against one of the native partner proteins to determine whether a protein of the predicted fusion size could be detected in cell lysates from cell lines with and without RNA transcript evidence of the fusion. We observed specific Western blots of the targeted protein at the expected canonical size and detected protein at the predicted fusion size specifically in the cell lines with the fusion transcripts, and not in cell lines without the fusions for both of the breast cancer associated read-through fusion transcripts (Fig. 3). The cell line with the most fusion-spanning reads was positive for the fusion in both Western blots, and in the case of the SCNN1A-TNFRSF1A, the cell line with the second highest number of fusion-spanning reads, was also positive by Western blot. These results suggest that the breast cancer associated read-through fusion transcripts are translated into fusion proteins. This observation raises the possibility that these cancer-specific fusion proteins may be expressed on the membrane of breast cancer cells and warrants further investigation as potential cell surface antibody drug targets.
Western blots of breast cancer associated fusion proteins. We performed Western blots using antibodies raised to one of the fusion partner proteins for the breast cancer associated fusion transcripts. For each candidate fusion, we ran cell lysates from two cell lines with RNA-seq reads spanning the fusion junction and one cell line without RNA-seq reads spanning the fusion junction. In each blot, the canonical/native size of the targeted protein was detected in each cell line, and a band at the predicted fusion protein size was detected in the cell line with the most RNA-seq fusion-spanning reads (CTSD-IFITM10 in MCF-7, and SCNN1A-TNFRSF1A in HCC1954). A band corresponding to the size of the predicted fusion protein was also detected in the cell line with the second most RNA-seq fusion transcript reads for the SCNN1A-TNFRSF1A fusion (SUM-102). None of the cell lines without RNA-seq evidence of the fusion transcript produced fusion protein-sized bands
Fusion transcript associated with proliferation
The CTSD-IFITM10 fusion transcript appears to be breast cancer specific, i.e., it was detected exclusively in breast cancer samples and not detected in any normal tissues. It was also detected in RNA-seq data from the MCF7 breast cancer cell line, which makes it amenable to further investigation in vitro. We designed two custom siRNA duplexes to target the fusion junction of the read-through fusion transcript (Fig. 4a). We transfected the MCF7 cell line with the siRNA duplexes targeting the fusion transcript and measured the abundance of fusion transcript 48 h after transfection using quantitative PCR (qPCR) with primers flanking the fusion junction (Fig. 4a). Both siRNAs targeting the fusion junction of CTSD-IFITM10 produced knockdown of the fusion transcript resulting in 42–51 % of the transcript remaining relative to treatment with a non-targeting siRNA (Fig. 4b). To determine if knockdown of the fusion transcript affects cell proliferation, we measured the number of live cells 72 h after transfection with each siRNA targeting the fusion junction. We found that both siRNAs targeting the CTSD-IFITM10 fusion transcript resulted in a significant decrease in the number of live cells (p < 0.03) resulting in 10–17 % reduction in live cell numbers compared to treatment with the non-targeting siRNA (Fig. 4c). While this decrease is modest, it is important to note that this cell phenotype is evident even when 45 % of the fusion transcript remains after knockdown. This qPCR detection and siRNA knockdown further confirm the presence of the CTSD-IFITM10 read-through fusion transcript and indicate that its abundance is associated with MCF7 breast cancer cell proliferation.
CTSD-IFITM10 read-through fusion transcript siRNA knockdown. a We designed qPCR primers to flank the fusion junction of the CTSD-IFITM10 read-through fusion transcript and we designed two custom siRNAs to target the fusion junction. The sequence from the CTSD (the 5′ gene) is indicated in green and the sequence from IFITM10 (3′ gene) is indicated in red. The MCF7 breast cancer cell line was transfected with two siRNAs targeting the CTSD-IFITM10 fusion junction. b qPCR of the fusion transcript was performed 48 h after transfection. Both siRNAs significantly reduced the abundance of the fusion transcript relative to the controls, which included a non-targeting siRNA and a mock transfection that did not contain any siRNA. c A quantitative cell proliferation assay was performed 72 h after transfection. Both siRNAs significantly reduced the number of live cells relative to the controls
Discussion
To our knowledge, this is the first report characterizing recurrent read-through fusion transcripts associated with breast cancer. Significant effort has been devoted to identifying gene expression differences and DNA mutations in breast cancer, and this report adds aberrant mRNA read-through fusion transcripts to the list of molecular defects associated with the disease. Both recurrent fusion transcripts associated with breast cancer involved membrane proteins, which raises the exciting possibility that they are breast cancer-specific cell surface markers that could be targeted with antibody–drug conjugates. In the MCF7 breast cancer cell line, the siRNA knockdown of CTSD-IFITM10 fusion was associated with a decrease in live cells suggesting this fusion plays a role in breast cancer cell proliferation. Read-through fusion transcripts represent a new class of exciting candidate biomarkers and potential therapeutic targets for further investigation in breast cancer. Future work to elucidate the mechanisms leading to the read-through transcription, mis-splicing, and loss of polyadenylation that create these fusions is also warranted to determine whether a defect in the regulation of these processes is responsible for these aberrant transcripts.
Materials and methods
Cell lines and tissues
De-identified fresh frozen breast cancer specimens, fresh frozen matched uninvolved breast tissue adjacent to tumors, and fresh frozen breast tissue specimens from reduction mammoplasty procedures were obtained from the University of Alabama at Birmingham’s Comprehensive Cancer Center Tissue Procurement Shared Facility. The specific aliquots of specimens provided for research were chosen based on their quality control by board certified pathologists. After identification by quality control, the uninvolved breast tissue aliquots were not further macro-dissected. The breast tumor specimens were macro-dissected by the pathologists at the Tissue Procurement Shared Facility to enrich for tumor cell content and remove adjacent normal tissue. The frozen breast tissue specimens were weighed, transferred to a 15 mL conical tube containing ceramic beads, and RLT Buffer (Qiagen) plus 1 % BME was added so that the tube contained 35 μL of buffer for each milligram of tissue. The conical tubes containing tissue, ceramic beads and buffer were then shaken in a MP Biomedicals FastPrep machine until the tissue was visibly homogenized (90 s at 6.5 meters per second). The homogenized tissue was stored at −80 °C. The 28 breast cancer cell lines were cultured as described previously [41].
RNA-seq
Total RNA was extracted from 5 million cultured cells or 350 μL of tissue homogenate (equivalent to 10 mg of tissue) using the Norgen Animal Tissue RNA Purification Kit (Norgen Biotek Corporation). Cell lysate was treated with Proteinase K before it was applied to the column, and on-column DNAse treatment was performed according to the manufacturer’s instructions. Total RNA was eluted from the columns and quantified using the Qubit RNA Assay Kit and the Qubit 2.0 fluorometer (Invitrogen). RNA-seq libraries for each sample were constructed from 250 ng total RNA using the polyA selection and transposase-based non-stranded library construction (Tn-RNA-seq) described previously [35]. RNA-seq libraries were barcoded during PCR using Nextera barcoded primers according to the manufacturer (Epicentre). The RNA-seq libraries were quantified using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Invitrogen), and three barcoded libraries were pooled in equimolar quantities for sequencing. The pooled libraries were sequenced on an Illumina HiSeq 2000 sequencing machine using paired-end 50 bp reads and a 6 bp index read, and we obtained at least 50 million read pairs from each library. ChimeraScan 0.4.5a was used to align reads to the hg19 human reference genome and utilize the UCSC Known Gene annotation file to identify fusion transcripts in each of the sequencing libraries [36]. ChimeraScan 0.4.5a default parameters were used, including using the bowtie -best -strata option for alignment, 2 mismatches tolerated at breakpoints, 4 bp minimum overlap required to call spanning reads, 8 bp anchor region where mismatch checks are enforced, and 0 mismatches allowed within the anchor region. Default filters include removing chimeras with less than 2 unique aligned fragments, removing chimeras when the probability of observing the putative insert size is than 0.01, or when the expression ratio relative to the wild-type transcripts is less than 0.01. To quantify the expression of each fusion partner gene, we used TopHat v1.4.1 [42] with the options -r 100 -mate-std-dev 75 to align 50 million RNA-seq read pairs, and used GENCODE version 9 [43] as a transcript reference. Gene expression values (fragments per kilobase of transcript per million reads, FPKMs) were calculated for each GENCODE transcript using Cufflinks 1.3.0 with the -u option [44].
Fusion transcript cDNA cloning and Sanger sequencing
Total RNA from the MCF-7 and HCC1954 breast cancer cell lines was extracted using the Norgen Animal Tissue RNA Purification Kit (Norgen Biotek Corporation). First strand cDNA was prepared from total RNA using Dynabeads oligo(dT) (Invitrogen) to select polyadenylated mRNA and SuperScript II Reverse Transcriptase with Random Hexamers (Invitrogen). PCR primers were designed to each exon in the fusion partner genes and used to amplify the SCNN1A-TNFRSF1A fusion transcript from HCC1954 and the CTSD-IFITM10 fusion transcript from MCF-7. PCR was performed using 0.5 µM each primer, 1 µL cDNA, 1× Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs), and 3 % DMSO. The largest PCR products were produced using the following primers: SCNN1A Forward (CTCTGCACCTTTGGCATGATGTACT), TNFRSF1A Reverse (GGACAGTTCAGCTTGCTATGTGCTT), CTSD Forward (ATGCAGCCCTCCAGCCTTCT), IFITM10 Reverse (ATAAGCCCTTCCTGCTAGGTGTCAG). The PCR products were extracted from agarose gels using the Qiagen Qiaquick Gel Extraction Kit and A-tailing was performed using 2.5 U Klenow Fragment (3′ → 5′ exo-) (New England Biolabs) and 450 µM dATP in a 55-μL reaction containing 1× NEBuffer 2 (New England Biolabs). The PCR products were ligated into the pGEMT Easy vector (Promega) and transformed into JM109 High Efficiency Competent Cells (Promega). Blue white screening was used to select transformed clones for overnight liquid culture and plasmid preparation using Wizard Plus SV Miniprep DNA Purification System (Promega). Plasmids were sequenced from both ends of the PCR product insert using M13 pUC Forward and Reverse primers on ABI 3730XL sequencers by MC Lab (San Francisco, CA).
Splice junction DNA sequencing
Genomic DNA was isolated from 12 breast cancer cell lines using 5 million cultured cells per cell line and the Qiagen DNeasy Kit. PCR amplification of 200 bp surrounding the terminal exon splice acceptor site that is skipped in the formation of the read-through fusion transcripts were performed in 50 μL reactions containing 5 ng genomic DNA, 0.5 µM Forward PCR primer, 0.5 µM Reverse PCR primer, 5 units Platinum Taq DNA Polymerase (Invitrogen), 1× PCR Buffer with 2 mM MgCl2, 0.5 mM each dNTP, and 0.5 M Betaine. These reactions were denatured at 98 °C for 1 min then thermocycled (30 cycles of 95 °C for 30 s and 62 °C for 3 min) and held at 4 °C. The PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter). The PCR products were quantified using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Invitrogen). Equimolar quantities of each of the eight PCR products were pooled into 12 pools, one for each cell line. Illumina sequencing libraries were prepared for each of the 12 pools of PCR products using Nextera according to the manufacturer’s instructions (Epicentre). The 12 libraries were quantified using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Invitrogen). Equimolar quantities of each library were pooled and diluted to 10 nM and sequenced using single-end 50 bp reads and a 6 base index read on the Illumina MiSeq sequencer. We obtained 6 million sequencing reads in total covering all 8 amplicons in each of the 12 breast cancer cell lines. Variants were identified by the GATK software on BaseSpace (Illumina), and BAM files were downloaded and inspected manually using IGV 2.0 [45].
Western blots
Breast cancer cell pellets containing 2.5 million cells were lysed by adding 100 μL RIPA Buffer (1× PBS, 1 % NP-40, 0.5 % sodium deoxycholate, 0.1 % SDS, and Roche protease inhibitor cocktail) and passing the solution through a 21-gauge needle. The lysed cells were then centrifuged at 16,000 rcf for 15 min at 4 °C, and the supernatant was collected, and protein was quantified using the Qubit Protein Assay Kit and the Qubit 2.0 fluorometer (Invitrogen). Twenty micrograms of protein extract was loaded into a 12 % SDS–polyacrylamide gel in 1× Tris/Glycine Buffer (BioRad). Magic Marker (Invitrogen) was used as a protein standard. The gel electrophoresis rig was partially immersed in an ice bath while it ran for 1.5 h at 125 V. Proteins were transferred to a nitrocellulose membrane using the iBlot system (Invitrogen) for 7 min at 20 V. The membranes were washed (1× PBS with 0.05 % Tween 20) and incubated in blocking buffer for 60 min (1× PBS with 0.05 % Tween 20 and 5 % w/v Instant Nonfat Dry Milk). The membranes were then incubated with primary antibody overnight at 4 °C (1× PBS with 0.05 % Tween 20, 1 % w/v Instant Nonfat Dry Milk, and 500 ng/mL primary antibody) followed by three 10 min washes (1x PBS with 0.05 % Tween 20). The following primary antibodies from Santa Cruz Biotechnology were used: CTSD sc-374381, and TNFRSF1A sc-8436. The membrane was then incubated with secondary antibody (1× PBS, 0.05 % Tween 20, 1 % Instant Nonfat Dry Milk, and a 1:4,000 dilution of horseradish peroxidase (HRP) conjugated goat anti-mouse secondary antibody (Thermo Scientific)). The membrane was then washed (1x PBS with 0.05 % Tween 20) and incubated for 5 min in a substrate solution of equal parts stable peroxide and luminol/enhancer (SuperSignal West Femto Chemiluminescent Substrate, Thermo Scientific). The membranes were then imaged for chemiluminescence.
Small interfering RNA (siRNA) knockdown
We ordered two ON-TARGETplus custom siRNA duplex reagents from Thermo Scientific that were designed to target the fusion junctions of the read-through fusion transcript and we also purchased ON-TARGETplus Non-targeting siRNA #1 (Thermo Scientific catalog # # D-001810-01-05), to serve as a control in our experiments. To design our custom siRNAs, we first entered the fusion junction nucleotide sequences into the siDESIGN Center on the Thermo Scientific website. The software was successfully designed CTSD-IFITM10 siRNA #1 to the fusion junction. The software did not report any other siRNAs. We then manually entered the fusion junction sequence for CTSD-IFITM10 siRNA #2, so that we would have a second siRNA targeting each fusion junction sequence with a more even representation of bases on each side of the junction. The siRNA duplex sequences are as follows:
CTSD-IFITM10 siRNA #1
Sense: ACUACACGCUCAAGGCCCAUU
Antisense: 5′P-UGGGCCUUGAGCGUGUAGUUU
CTSD-IFITM10 siRNA #2
Sense: ACGCUCAAGGCCCAGGGCCUU
Antisense: 5′-PGGCCCUGGGCCUUGAGCGUUU
The siRNA transfection experiments were performed in 96-well plates in triplicate, and included a mock transfection control with no siRNA, a non-targeting siRNA control, and the two custom siRNAs targeting the fusion junction. The Lipofectamine RNAiMAX Transfection Reagent and siRNA were prepared according the manufacturer’s instructions (Invitrogen). We added 10 µL of the mix containing siRNA and transfection reagent diluted in Opti-MEM I Reduced Serum Medium (Invitrogen) to each well in the 96-well plate containing cells, which results in 3 pmol of siRNA in 0.3 μL of Lipofectamine RNAiMAX reagent per well.
Quantitative PCR (qPCR)
We ordered PCR primers flanking the fusion junction of the CTSD-IFITM10 read-through fusion transcript, as well as primers to the CTCF gene, which were used as a control for normalization. The primer oligonucleotide sequences are as follows:
CTSD-IFITM10 qPCR Primers:
Forward: CTACAAGCTGTCCCCAGAGG
Reverse: CCGTCCGTGGTGCTG
CTCF qPCR Primers:
Forward: ACCTGTTCCTGTGACTGTACC
Reverse: ATGGGTTCACTTTCCGCAAGG
For siRNA experiments, we performed the qPCR assay 48 h after transfection. We prepared cDNA using the Power SYBR Green Cells-to-CT Kit (Invitrogen) according to the manufacturer’s instructions, including the option of using 22.5 μL of cell lysate in the reverse transcription reaction. For normal breast tissue experiments, we prepared cDNA from 10 ng total RNA using the SuperScript II (Invitrogen) Reverse Transcription Kit according to the manufacturer’s instruction. Normal tissue cDNA was diluted with 60 µL of water before qPCR.
qPCR experiments were run in duplicate in 10 μL reactions with 4 μL of cDNA, 5 μL Power SYBR Green PCR Master Mix, and PCR primers added to a final concentration of 200 nM. For each cDNA sample, we also performed control qPCR experiments using 400 nM of each primer designed to CTCF, a housekeeping gene locus that we used to ensure that the quantity and quality of cDNA were equivalent across experiments. The reactions were run on an ABI 7900HT with the following thermal cycling conditions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. A dissociation curve analysis was run using the standard protocol on the instrument. Transcript abundance was calculated using automatic baseline and threshold settings using the instrument’s software. To calculate the percentage of transcript remaining after siRNA knockdown, we first computed the fusion transcript delta cycle threshold (dCt) by normalizing the fusion transcript abundance measured in wells treated with siRNAs targeting the fusion junction to the transcript abundance measured in wells treated with the non-targeting siRNA. We then calculated the dCt values of the CTCF housekeeping control locus from the same samples. We subtracted the CTCF dCt from the fusion transcript dCt to compute the ddCt values and compute the fold change of the fusion transcript expression. As an additional control, we also performed this ddCt calculation on the mock transfection with no siRNA to ensure that the presence of the non-targeting siRNA did not affect the abundance of the fusion transcript.
Cell proliferation
We performed cell proliferation assays 72 h after transfection using the CyQUANT Cell Proliferation Assay Kit for Cells in Culture (Invitrogen) according to the manufacturer‘s instruction. Our protocol included using 1.5× CyQUANT GR dye, which was recommended to obtain adequate dynamic range in wells with 75,000 cells. The fluorescence from each well of the 96-well plate was measured using the Molecular Devices SpectraMax M5e plate reader. To calculate the percentage of live cells remaining after siRNA knockdown, we normalized the fluorescence intensity in wells treated with siRNAs targeting the fusion junction to the fluorescence measured in wells treated with the non-targeting siRNA. As a control, we also performed this normalization on the mock transfection with no siRNA to ensure that the presence of the non-targeting siRNA did not affect the fluorescence or quantity of live of the cells.
Data access
All RNA-seq data generated in this study are available for download from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) through accession number GSE58135.
References
Nowell PC (1962) The minute chromosome (Phl) in chronic granulocytic leukemia. Blut 8:65–66
Rowley JD (1973) Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243(5405):290–293
Rowley JD (2008) Chromosomal translocations: revisited yet again. Blood 112(6):2183–2189
Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB (1996) Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr–Abl positive cells. Nat Med 2(5):561–566
Mitelman F, Johansson B, Mertens F (2004) Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet 36(4):331–334
Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, Lonigro RJ, Schroth G, Kumar-Sinha C, Chinnaiyan AM (2009) Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA 106(30):12353–12358
Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH (2002) Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2(5):367–376
Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G (2009) Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA 106(44):18740–18744
Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM (2007) Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448(7153):595–599
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310(5748):644–648
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM (2008) Recurrent gene fusions in prostate cancer. Nat Rev Cancer 8(7):497–511
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H (2007) Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448(7153):561–566
Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, Lonigro RJ, Quist M, Siddiqui J, Mehra R, Jing X, Giordano TJ, Sabel MS, Kleer CG, Palanisamy N, Natrajan R, Lambros MB, Reis-Filho JS, Kumar-Sinha C, Chinnaiyan AM (2011) Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 17(12):1646–1651
Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, Sun Z, Chai HS, Williamson DW, Radisky D, Schroth GP, Kocher JP, Perez EA, Thompson EA (2011) A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res 39(15):e100
Asmann YW, Necela BM, Kalari KR, Hossain A, Baker TR, Carr JM, Davis C, Getz JE, Hostetter G, Li X, McLaughlin SA, Radisky DC, Schroth GP, Cunliffe HE, Perez EA, Thompson EA (2012) Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res 72(8):1921–1928
Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, Rye IH, Nyberg S, Wolf M, Borresen-Dale AL, Kallioniemi O (2011) Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 12(1):R6
Ha KC, Lalonde E, Li L, Cavallone L, Natrajan R, Lambros MB, Mitsopoulos C, Hakas J, Kozarewa I, Fenwick K, Lord CJ, Ashworth A, Vincent-Salomon A, Basik M, Reis-Filho JS, Majewski J, Foulkes WD (2011) Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genomics 4:75
Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, Rogers YH, Venter JC, Simpson AJ, Strausberg RL (2009) Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci USA 106(6):1886–1891
Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12(8):R72
Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, Del Pozo A, Tress M, Johnson R, Guigo R, Valencia A (2012) Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res 22(7):1231–1242
Li X, Zhao L, Jiang H, Wang W (2009) Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J Mol Evol 68(1):56–65
Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R (2006) Transcription-mediated gene fusion in the human genome. Genome Res 16(1):30–36
Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R (2006) Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res 16(1):37–44
Li H, Wang J, Ma X, Sklar J (2009) Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle 8(2):218–222
Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, Rubin MA (2009) SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 69(7):2734–2738
Kim RN, Kim A, Choi SH, Kim DS, Nam SH, Kim DW, Kang A, Kim MY, Park KH, Yoon BH, Lee KS, Park HS (2012) Novel mechanism of conjoined gene formation in the human genome. Funct Integr Genomics 12(1):45–61
Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, Nishida Y, Fujikake T, Takeda T, Taylor TD (2010) Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLoS One 5(10):e13284
Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM (2012) SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing. Cancer Discov 2(7):582–585
Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, Stinson J, Peters BA, Modrusan Z, Jung K, Seshagiri S, Wu TD (2011) Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics 4:11
Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM (2009) Transcriptome sequencing to detect gene fusions in cancer. Nature 458(7234):97–101
Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H (2012) Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov 2(7):598–607
Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L (2011) Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA 108(22):9172–9177
Zhou J, Liao J, Zheng X, Shen H (2012) Chimeric RNAs as potential biomarkers for tumor diagnosis. BMB Rep 45(3):133–140
Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R, Sieuwerts AM, Martens JW, Silver DP, Langerod A, Russnes HE, Foekens JA, Reis-Filho JS, van ‘t Veer L, Richardson AL, Borresen-Dale AL, Campbell PJ, Futreal PA, Stratton MR (2009) Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462(7276):1005–1010
Gertz J, Varley KE, Davis NS, Baas BJ, Goryshin IY, Vaidyanathan R, Kuersten S, Myers RM (2012) Transposase mediated construction of RNA-seq libraries. Genome Res 22(1):134–141
Iyer MK, Chinnaiyan AM, Maher CA (2011) ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 27(20):2903–2904
Hummler E, Beermann F (2000) Scnn1 sodium channel gene family in genetically engineered mice. J Am Soc Nephrol 11(Suppl 16):S129–S134
Chen G, Goeddel DV (2002) TNF-R1 signaling: a beautiful pathway. Science 296(5573):1634–1635
Nicotra G, Castino R, Follo C, Peracchio C, Valente G, Isidoro C (2010) The dilemma: does tissue expression of cathepsin D reflect tumor malignancy? The question: does the assay truly mirror cathepsin D mis-function in the tumor? Cancer Biomark 7(1):47–64
Hickford D, Frankenberg S, Shaw G, Renfree MB (2012) Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genom 13:155
Oliver PG, LoBuglio AF, Zhou T, Forero A, Kim H, Zinn KR, Zhai G, Li Y, Lee CH, Buchsbaum DJ (2012) Effect of anti-DR5 and chemotherapy on basal-like breast cancer. Breast Cancer Res Treat 133(2):417–426
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R (2006) GENCODE: producing a reference annotation for ENCODE. Genome Biol 7(Suppl 1):S4 1–9
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26
Acknowledgements
This study was supported in part by funding from TATRC, USAMRMC (W81XWH1010790), a Komen for the Cure Promise Grant (KG090969), and the National Institutes of Health, The National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (P50CA089019).
Conflict of interest
The authors do not have a financial relationship with the organizations that sponsored this research. Following the preparation of the manuscript the HudsonAlpha Institute for Biotechnology has filed a patent application describing the discovery and detection of the fusion transcripts in breast cancer. A research collaboration between the HudsonAlpha Institute for Biotechnology and Seattle Genetics, Inc. has been established to investigate whether these fusion transcripts could be therapeutic targets. The manuscript authors (RMM, KEV, BSR, JG, DJB, AF, AFL) are inventors on the patent and could receive royalties if the eventual commercialization of a drug involves licensing the patent.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Varley, K.E., Gertz, J., Roberts, B.S. et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat 146, 287–297 (2014). https://doi.org/10.1007/s10549-014-3019-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3019-2