Abstract
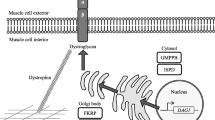
Duchenne/Becker muscular dystrophies are the most frequent inherited neuromuscular diseases caused by mutations of the dystrophin gene. However, approximately 30 % of patients with the disease do not receive a molecular diagnosis because of the complex mutational spectrum and the large size of the gene. The introduction and use of next-generation sequencing have advanced clinical genetic research and might be a suitable method for the detection of various types of mutations in the dystrophin gene. To identify the mutational spectrum using a single platform, whole dystrophin gene sequencing was performed using next-generation sequencing. The entire dystrophin gene, including all exons, introns and promoter regions, was target enriched using a DMD whole gene enrichment kit. The enrichment libraries were sequenced on an Illumina HiSeq 2000 sequencer using paired read 100 bp sequencing. We studied 26 patients: 21 had known large deletion/duplications and 5 did not have detectable large deletion/duplications by multiplex ligation-dependent probe amplification technology (MLPA). We applied whole dystrophin gene analysis by next-generation sequencing to the five patients who did not have detectable large deletion/duplications and to five randomly chosen patients from the 21 who did have large deletion/duplications. The sequencing data covered almost 100 % of the exonic region of the dystrophin gene by ≥10 reads with a mean read depth of 147. Five small mutations were identified in the first five patients, of which four variants were unreported in the dmd.nl database. The deleted or duplicated exons and the breakpoints in the five large deletion/duplication patients were precisely identified. Whole dystrophin gene sequencing by next-generation sequencing may be a useful tool for the genetic diagnosis of Duchenne and Becker muscular dystrophies.


Similar content being viewed by others
References
Bennett RR, den Dunnen J, O’Brien KF, Darras BT, Kunkel LM (2001) Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet 2:17
Bovolenta M, Neri M, Fini S, Fabris M, Trabanelli C, Venturoli A, Martoni E, Bassi E, Spitali P, Brioschi S, Falzarano MS, Rimessi P, Ciccone R, Ashton E, McCauley J, Yau S, Abbs S, Muntoni F, Merlini L, Gualandi F, Ferlini A (2008) A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies. BMC Genom 9:572
Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT (1988) Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16:11141–11156
Chen WJ, Lin QF, Zhang QJ, He J, Liu XY, Lin MT, Murong SX, Liou CW, Wang N (2013) Molecular analysis of the dystrophin gene in 407 Chinese patients with Duchenne/Becker muscular dystrophy by the combination of multiplex ligation-dependent probe amplification and Sanger sequencing. Clin Chim Acta 423C:35–38
Deburgrave N, Daoud F, Llense S, Barbot JC, Recan D, Peccate C, Burghes AH, Beroud C, Garcia L, Kaplan JC, Chelly J, Leturcq F (2007) Protein- and mRNA-based phenotype–genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum Mutat 28:183–195
Dent KM, Dunn DM, von Niederhausern AC, Aoyagi AT, Kerr L, Bromberg MB, Hart KJ, Tuohy T, White S, den Dunnen JT, Weiss RB, Flanigan KM (2005) Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am J Med Genet A 134:295–298
Dolinsky LC, de Moura-Neto RS, Falcao-Conceicao DN (2002) DGGE analysis as a tool to identify point mutations, de novo mutations and carriers of the dystrophin gene. Neuromuscul Disord 12:845–848
Dwi Pramono ZA, Takeshima Y, Surono A, Ishida T, Matsuo M (2000) A novel cryptic exon in intron 2 of the human dystrophin gene evolved from an intron by acquiring consensus sequences for splicing at different stages of anthropoid evolution. Biochem Biophys Res Commun 267:321–328
Emery AE (2002) The muscular dystrophies. Lancet 359:687–695
Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB (2003) Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet 72:931–939
Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM, Pestronk A, Florence JM, Connolly AM, Mathews KD, Stephan CM, Laubenthal KS, Wong BL, Morehart PJ, Meyer A, Finkel RS, Bonnemann CG, Medne L, Day JW, Dalton JC, Margolis MK, Hinton VJ, Weiss RB (2009) Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 30:1657–1666
Gatta V, Scarciolla O, Gaspari AR, Palka C, De Angelis MV, Di Muzio A, Guanciali-Franchi P, Calabrese G, Uncini A, Stuppia L (2005) Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiple ligation probe amplification (MLPA). Hum Genet 117:92–98
Grimm T, Kress W, Meng G, Muller CR (2012) Risk assessment and genetic counseling in families with Duchenne muscular dystrophy. Acta Myol 31:179–183
Hamed SA, Hoffman EP (2006) Automated sequence screening of the entire dystrophin cDNA in Duchenne dystrophy: point mutation detection. Am J Med Genet B Neuropsychiatr Genet 141B:44–50
Helderman-van den Enden AT, Straathof CS, Aartsma-Rus A, den Dunnen JT, Verbist BM, Bakker E, Verschuuren JJ, Ginjaar HB (2010) Becker muscular dystrophy patients with deletions around exon 51; a promising outlook for exon skipping therapy in Duchenne patients. Neuromuscul Disord 20:251–254
Hofstra RM, Mulder IM, Vossen R, de Koning-Gans PA, Kraak M, Ginjaar IB, van der Hout AH, Bakker E, Buys CH, van Ommen GJ, van Essen AJ, den Dunnen JT (2004) DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients. Hum Mutat 23:57–66
Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D, Djurisic M, Breuning MH, White SJ, den Dunnen JT (2005) Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet 13:1231–1234
Lenk U, Oexle K, Voit T, Ancker U, Hellner KA, Speer A, Hubner C (1996) A cysteine 3340 substitution in the dystroglycan-binding domain of dystrophin associated with Duchenne muscular dystrophy, mental retardation and absence of the ERG b-wave. Hum Mol Genet 5:973–975
Lim BC, Lee S, Shin JY, Kim JI, Hwang H, Kim KJ, Hwang YS, Seo JS, Chae JH (2011) Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: comprehensive mutational search in a single platform. J Med Genet 48:731–736
Madania A, Zarzour H, Jarjour RA, Ghoury I (2010) Combination of conventional multiplex PCR and quantitative real-time PCR detects large rearrangements in the dystrophin gene in 59% of Syrian DMD/BMD patients. Clin Biochem 43:836–842
Marquis-Nicholson R, Lai D, Lan CC, Love JM, Love DR (2013) A streamlined protocol for molecular testing of the DMD Gene within a Diagnostic Laboratory: a combination of array comparative genomic hybridization and bidirectional sequence analysis. ISRN Neurol 2013:908317
Mendell JR, Buzin CH, Feng J, Yan J, Serrano C, Sangani DS, Wall C, Prior TW, Sommer SS (2001) Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology 57:645–650
Mulle JG, Patel VC, Warren ST, Hegde MR, Cutler DJ, Zwick ME (2010) Empirical evaluation of oligonucleotide probe selection for DNA microarrays. PLoS ONE 5:e9921
Murugan S, Chandramohan A, Lakshmi BR (2010) Use of multiplex ligation-dependent probe amplification (MLPA) for Duchenne muscular dystrophy (DMD) gene mutation analysis. Indian J Med Res 132:303–311
Oudet C, Hanauer A, Clemens P, Caskey T, Mandel JL (1992) Two hot spots of recombination in the DMD gene correlate with the deletion prone regions. Hum Mol Genet 1:599–603
Roberts RG, Bentley DR, Bobrow M (1993) Infidelity in the structure of ectopic transcripts: a novel exon in lymphocyte dystrophin transcripts. Hum Mutat 2:293–299
Sansovic I, Barisic I, Dumic K (2013) Improved detection of deletions and duplications in the DMD gene using the multiplex ligation-dependent probe amplification (MLPA) method. Biochem Genet 51:189–201
Shyr D, Liu Q (2013) Next generation sequencing in cancer research and clinical application. Biol Proced Online 15:4
Suminaga R, Takeshima Y, Adachi K, Yagi M, Nakamura H, Matsuo M (2002) A novel cryptic exon in intron 3 of the dystrophin gene was incorporated into dystrophin mRNA with a single nucleotide deletion in exon 5. J Hum Genet 47:196–201
Sun H, Chasin LA (2000) Multiple splicing defects in an intronic false exon. Mol Cell Biol 20:6414–6425
Sura T, Eu-ahsunthornwattana J, Pingsuthiwong S, Busabaratana M (2008) Sensitivity and frequencies of dystrophin gene mutations in Thai DMD/BMD patients as detected by multiplex PCR. Dis Markers 25:115–121
Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M (1999) Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 8:493–500
Takeshima Y, Yagi M, Okizuka Y, Awano H, Zhang Z, Yamauchi Y, Nishio H, Matsuo M (2010) Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet 55:379–388
Todorova A, Todorov T, Georgieva B, Lukova M, Guergueltcheva V, Kremensky I, Mitev V (2008) MLPA analysis/complete sequencing of the DMD gene in a group of Bulgarian Duchenne/Becker muscular dystrophy patients. Neuromuscul Disord 18:667–670
Tran VK, Zhang Z, Yagi M, Nishiyama A, Habara Y, Takeshima Y, Matsuo M (2005) A novel cryptic exon identified in the 3′ region of intron 2 of the human dystrophin gene. J Hum Genet 50:425–433
Traverso M, Malnati M, Minetti C, Regis S, Tedeschi S, Pedemonte M, Bruno C, Biassoni R, Zara F (2006) Multiplex real-time PCR for detection of deletions and duplications in dystrophin gene. Biochem Biophys Res Commun 339:145–150
Trimarco A, Torella A, Piluso G, Maria Ventriglia V, Politano L, Nigro V (2008) Log-PCR: a new tool for immediate and cost-effective diagnosis of up to 85 % of dystrophin gene mutations. Clin Chem 54:973–981
White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebregt E, Bakker B, van Ommen GJ, Breuning MH, den Dunnen JT (2002) Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet 71:365–374
White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers AL, Lalic T, Janson AA, Ginjaar HB, Breuning MH, den Dunnen JT (2006) Duplications in the DMD gene. Hum Mutat 27:938–945
Xie S, Lan Z, Qu N, Wei X, Yu P, Zhu Q, Yang G, Wang J, Shi Q, Wang W, Yang L, Yi X (2012) Detection of truncated dystrophin lacking the C-terminal domain in a Chinese pedigree by next-generation sequencing. Gene 499:139–142
Yan J, Feng J, Buzin CH, Scaringe W, Liu Q, Mendell JR, den Dunnen J, Sommer SS (2004) Three-tiered noninvasive diagnosis in 96% of patients with Duchenne muscular dystrophy (DMD). Hum Mutat 23:203–204
Yang J, Li SY, Li YQ, Cao JQ, Feng SW, Wang YY, Zhan YX, Yu CS, Chen F, Li J, Sun XF, Zhang C (2013) MLPA-based genotype–phenotype analysis in 1053 Chinese patients with DMD/BMD. BMC Med Genet 14:29
Zeng F, Ren ZR, Huang SZ, Kalf M, Mommersteeg M, Smit M, White S, Jin CL, Xu M, Zhou DW, Yan JB, Chen MJ, van Beuningen R, den Dunnen J, Zeng YT, Wu Y (2008) Array-MLPA: comprehensive detection of deletions and duplications and its application to DMD patients. Hum Mutat 29:190–197
Zhang Z, Habara Y, Nishiyama A, Oyazato Y, Yagi M, Takeshima Y, Matsuo M (2007) Identification of seven novel cryptic exons embedded in the dystrophin gene and characterization of 14 cryptic dystrophin exons. J Hum Genet 52:607–617
Acknowledgments
We thank all the tested individuals, their families and collaborating clinicians for their participation. This work was supported by the National Natural Science Foundation of China (81300527), the National Science and Technology Support Program of China (2013BA112B00) and Beijing Medical Capital Development Foundation 2012. This study has neither been presented nor submitted or accepted anywhere.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Y. Wang, Y. Yang, and J. Liu contributed equally to the paper.
Rights and permissions
About this article
Cite this article
Wang, Y., Yang, Y., Liu, J. et al. Whole dystrophin gene analysis by next-generation sequencing: a comprehensive genetic diagnosis of Duchenne and Becker muscular dystrophy. Mol Genet Genomics 289, 1013–1021 (2014). https://doi.org/10.1007/s00438-014-0847-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0847-z