Abstract
Noonan syndrome is a developmental disorder characterized by short stature, facial dysmorphia, congenital heart defects and skeletal anomalies1. Increased RAS-mitogen-activated protein kinase (MAPK) signaling due to PTPN11 and KRAS mutations causes 50% of cases of Noonan syndrome2,3,4,5,6. Here, we report that 22 of 129 individuals with Noonan syndrome without PTPN11 or KRAS mutation have missense mutations in SOS1, which encodes a RAS-specific guanine nucleotide exchange factor. SOS1 mutations cluster at codons encoding residues implicated in the maintenance of SOS1 in its autoinhibited form. In addition, ectopic expression of two Noonan syndrome–associated mutants induces enhanced RAS and ERK activation. The phenotype associated with SOS1 defects lies within the Noonan syndrome spectrum but is distinctive, with a high prevalence of ectodermal abnormalities but generally normal development and linear growth. Our findings implicate gain-of-function mutations in a RAS guanine nucleotide exchange factor in disease for the first time and define a new mechanism by which upregulation of the RAS pathway can profoundly change human development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
13 December 2006
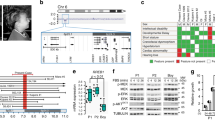
In the version of this article initially published online, the labels ‘expression’ in Figure 2 panels a, b and d are incorrect. The correct labels are ‘activation’. In addition, author Giuseppe Zampino should have affiliation 8 rather than affiliation 7. These errors have been corrected for all version of the article.
References
Noonan, J.A. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am. J. Dis. Child. 116, 373–380 (1968).
Carta, C. et al. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 79, 129–135 (2006).
Fragale, A., Tartaglia, M., Wu, J. & Gelb, B.D. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat. 23, 267–277 (2004).
Schubbert, S. et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331–336 (2006).
Tartaglia, M. et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am. J. Hum. Genet. 70, 1555–1563 (2002).
Tartaglia, M. et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468 (2001).
Keilhack, H., David, F.S., McGregor, M., Cantley, L.C. & Neel, B.G. Diverse biochemical properties of Shp2 mutants: implications for disease phenotypes. J. Biol. Chem. 280, 30984–30993 (2005).
Tartaglia, M. et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 78, 279–290 (2006).
Digilio, M.C. et al. Grouping of multiple-Lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 71, 389–394 (2002).
Legius, E. et al. PTPN11 mutations in LEOPARD syndrome. J. Med. Genet. 39, 571–574 (2002).
Aoki, Y. et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37, 1038–1040 (2005).
Niihori, T. et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 38, 294–296 (2006).
Rodriguez-Viciana, P. et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311, 1287–1290 (2006).
Nimnual, A. & Bar-Sagi, D. The two hats of SOS. Sci. STKE 2002, PE36 (2002).
Corbalan-Garcia, S., Margarit, S.M., Galron, D., Yang, S.S. & Bar-Sagi, D. Regulation of Sos activity by intramolecular interactions. Mol. Cell. Biol. 18, 880–886 (1998).
Sondermann, H., Nagar, B., Bar-Sagi, D. & Kuriyan, J. Computational docking and solution x-ray scattering predict a membrane-interacting role for the histone domain of the Ras activator son of sevenless. Proc. Natl. Acad. Sci. USA 102, 16632–16637 (2005).
Sondermann, H. et al. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 (2004).
Margarit, S.M. et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003).
Hart, T.C. et al. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am. J. Hum. Genet. 70, 943–954 (2002).
Qian, X. et al. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 19, 642–654 (2000).
Wang, D.Z. et al. Mutation in Sos1 dominantly enhances a weak allele of the EGFR, demonstrating a requirement for Sos1 in EGFR signaling and development. Genes Dev. 11, 309–320 (1997).
Sibilia, M. et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102, 211–220 (2000).
Vojtek, A.B. & Der, C.J. Increasing complexity of the Ras signaling pathway. J. Biol. Chem. 273, 19925–19928 (1998).
Nimnual, A.S., Yatsula, B.A. & Bar-Sagi, D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279, 560–563 (1998).
Bowtell, D., Fu, P., Simon, M. & Senior, P. Identification of murine homologues of the Drosophila son of sevenless gene: potential activators of ras. Proc. Natl. Acad. Sci. USA 89, 6511–6515 (1992).
Yang, S.S., Van Aelst, L. & Bar-Sagi, D. Differential interactions of human Sos1 and Sos2 with Grb2. J. Biol. Chem. 270, 18212–18215 (1995).
Collins, J.S. & Schwartz, C.E. Detecting polymorphisms and mutations in candidate genes. Am. J. Hum. Genet. 71, 1251–1252 (2002).
Sharland, M., Burch, M., McKenna, W.M. & Paton, M.A. A clinical study of Noonan syndrome. Arch. Dis. Child. 67, 178–183 (1992).
Acknowledgements
We are indebted to the individuals and families who participated in the study, the physicians who referred the subjects and the Joint Genome Institute Production Sequencing Group. We thank J. Kuriyan and H. Sondermann for helpful discussions. This work was supported by Telethon-Italy (grant GGP04172) and the 'Programma di Collaborazione Italia-USA/Malattie Rare' (M.T.); US National Institutes of Health grants HL71207, HD01294 and HL074728 (B.D.G.) and CA55360 and CA28146 (D.B.-S.) and Italian Ministry of Health Grant RC 2006 (B.D.). Research conducted at the E.O. Lawrence Berkeley National Laboratory and the Joint Genome Institute was performed under Department of Energy contract DE-AC0378SF00098 at the University of California (L.A.P.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
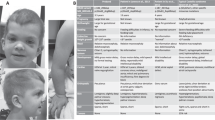
Facial dysmorphia in SOS1-associated Noonan syndrome. (PDF 288 kb)
Supplementary Table 1
Genotype-phenotype correlation (PDF 70 kb)
Rights and permissions
About this article
Cite this article
Tartaglia, M., Pennacchio, L., Zhao, C. et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet 39, 75–79 (2007). https://doi.org/10.1038/ng1939
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1939
This article is cited by
-
Novel effects of Ras-MAPK pathogenic variants on the developing human brain and their link to gene expression and inhibition abilities
Translational Psychiatry (2023)
-
An Assessment of the Therapeutic Landscape for the Treatment of Heart Disease in the RASopathies
Cardiovascular Drugs and Therapy (2023)
-
Cutis verticis gyrata and Noonan syndrome: report of two cases with pathogenetic variant in SOS1 gene
Italian Journal of Pediatrics (2022)
-
MR lymphangiography of lymphatic abnormalities in children and adults with Noonan syndrome
Scientific Reports (2022)
-
Altered canonical and striatal-frontal resting state functional connectivity in children with pathogenic variants in the Ras/mitogen-activated protein kinase pathway
Molecular Psychiatry (2022)