Article Text
Abstract
The study of patients with rare multiple congenital anomaly syndromes can provide illuminating insights into normal development and the pathogenesis of congenital anomalies. The GLI3 gene is a particularly good example as it illuminates the phenomena of pleiotropy, phenocopies, syndrome families, and evolutionary conservation of pathogenesis, and raises questions about how diagnoses are conceptualised. These topics are reviewed in turn, in the context of the clinical and biological data derived from patients with mutations in GLI3 and experimental work in model systems.
- ACLS, acrocallosal syndrome
- GCPS, Greig cephalopolysyndactyly syndrome
- PHS, Pallister-Hall syndrome
- modifier
- pleiotropy
- polydactyly
- transcription factor
- variable expressivity
Statistics from Altmetric.com
Interruption of the GLI3 gene (OMIM 165240) by translocations in 7p13 was originally described in 1991 in a small series of patients with the Greig cephalopolysyndactyly syndrome (GCPS, OMIM 175700),1 which is inherited in an autosomal dominant pattern. This syndrome comprises the dyad of polydactyly and craniofacial anomalies2 (table 1). Although the polydactyly is classically described as preaxial, the more common presentation is preaxial polydactyly of the feet and postaxial polydactyly of the hands (crossed polydactyly, type I, OMIM 174700).3 The craniofacial anomalies of GCPS include macrocephaly and hypertelorism with a broad nasal bridge. Early reports of the disorder included craniosynostosis as a manifestation, but it has subsequently become clear that this is an uncommon manifestation of the disorder.
Features of Greig cephalopolysyndactyly syndrome and overlapping syndromes
After the molecular delineation of GCPS, linkage analysis of two families with autosomal dominant Pallister-Hall syndrome (PHS, OMIM 146510) was undertaken.4 The phenotype of PHS includes central or postaxial polydactyly, hypothalamic hamartoma, bifid epiglottis or laryngeal cleft, and pulmonary segmentation anomalies, and this disorder is also inherited in an autosomal dominant pattern. This phenotype thus had very little clinical overlap with GCPS. In spite of that lack of phenotypic overlap, PHS mapped to 7p13 and was eventually shown to be allelic to GCPS, with several GLI3 mutations described in patients with PHS.5
EVOLUTIONARY CONSERVATION ILLUMINATES GLI3 FUNCTION
The GLI3 protein is a zinc finger transcription factor that is expressed early in development. This transcription factor regulates downstream genes by direct binding to specific sequences in the promoter region of target genes.6 The GLI3 protein is a downstream mediator of the sonic hedgehog pathway, and this pathway includes several genes that cause abnormal phenotypes in the human when mutated (for example, SHH, PTC1, and CBP).7
The understanding of the pathogenesis of GLI3 was markedly facilitated by analyses of model organisms and comparing the model organism data with the human data. The bifunctional nature of GLI3 was a hypothesis based on two observations: the biological function of cubitus interruptus (ci, the homologue of the GLI gene family in Drosophila) and the position of truncation mutations in GLI3 in humans. The key biological insight was made by the Kornberg group, when they showed that ci, which normally localises to the cytoplasm, is under the regulation of hedgehog (hh, the fly homologue of the vertebrate hedgehog gene family, sonic, Indian, and desert hedgehog).8,9 In the presence of hh, the 155 kDa ci protein was translocated to the nucleus and activated downstream genes, whereas in the absence of hh, ci was proteolytically processed to an N-terminal 75 kDa form that repressed downstream genes. The human genetics insight was provided by the observation that mutations in GLI3 that cause Pallister-Hall syndrome are frameshift or nonsense truncating mutations and that these mutations occur in a particular region of the protein.5,10 Intriguingly, when one aligns the human GLI3 and the fly ci proteins, it appears that the human truncation mutations that cause PHS occur exclusively in the domains of the protein that are carboxy-terminal of the predicted protein processing cleavage point of the ci gene. This suggested that PHS truncation mutations perturb the SHH related regulation of GLI3 processing and that GLI3 is bifunctional, as is ci. Subsequent data have shown that this is correct,11 and that PHS truncation mutations generate a constitutive gain of function mutation of a GLI3 repressor protein that is likely to be independent of SHH controlled post-translational regulation.
Thus GLI3 is a component of an evolutionarily conserved developmental cassette or module that is used to accomplish various tasks throughout the developing organism. For example, the SHH/GLI pathway is used to specify the neural tube, craniofacial structures, the limb, the lung, and many others. This developmental module is generally used to specify positional or polarity information within part of the developing embryo. A well known example of this is in the developing limb bud where SHH functions as a morphogen in the zone of polarising activity to establish anterior-posterior polarity. In this pathway, SHH functions in a cell autonomous fashion through a cell surface receptor complex of patched (PTC1) and smoothened (SMO) (reviewed by Villavicencio et al7). As is true for many developmental cassette pathways, SHH triggers a derepression of a negative modulation of the pathway. That is, binding of SHH to PTC1 releases the negative interaction of PTC1 on SMO, which normally negatively modulates the downstream pathway members. The net effect of SHH binding to the cell surface complex is an activation of the pathway. Downstream from this complex, the cassette includes a complex of proteins that are tethered to a cytoplasmic complex and this includes GLI3. It is at this point that that the post-translational regulation of GLI3 occurs (as described above).
Thus GLI3 functions as a bifunctional mediator of the SHH pathway, either activating or repressing the transcription of downstream genes. This is in contrast to most transcription factors, which function as either activators or repressors (but not both). The modulatory function of most other transcription factors is attributable either to their presence or absence, but they are not bifunctional. This means that GLI3 has multiple distinct biological functions and those functions are related to the distinct phenotypes that are caused by different classes of mutations in the gene. Thus distantly related lines of research (Drosophila basic biology and clinical molecular genetics) have converged to provide a coherent pathogenic basis of two distinct but allelic human malformation syndromes. These studies show the value of evolutionary analysis of gene function and the study of model organisms to illuminate mechanisms of disease in humans.
PLEIOTROPY
That GLI3 should have the attribute of pleiotropy is unsurprising, once its functions are appreciated. It turns out that the pleiotropy of GLI3 operates on several levels. As described above, GLI3 functions in various organ systems at different times in development. For most gene products, this is the sole basis of pleiotropy. That a gene product like GLI3 specifies the dorsal-ventral polarity of the developing neural tube and the anterior-posterior polarity of the limb bud is sufficient to explain why Pallister-Hall syndrome includes polydactyly and structural CNS anomalies (for example, hypothalamic hamartoma). But the evaluation of the genotype–phenotype correlation of GLI3 revealed another level of correlation with the biological function of GLI3. This provided insight into biology and also serves as an example of the value of detailed clinical analysis of patients with rare disorders.
The finding that PHS was allelic to GCPS was unexpected and surprising. The anomalies of PHS overlap only minimally with GCPS and in fact the differential diagnosis of each of these two conditions includes several conditions, but not each other. For example, PHS has substantial clinical overlap with Bardet-Biedl syndrome, McKusick-Kaufmann syndrome, and several forms of the oral-facial-digital syndromes. In contrast, GCPS can be difficult to distinguish from frontonasal dysplasia spectrum, Opitz trigonencephaly syndrome, and others. It is ironic that PHS is easy to distinguish from GCPS in the clinic (see table 1). Part of the challenge with the differential of GCPS is that the cardinal features of that disorder are relatively non-specific. Preaxial polydactyly is recognised in more than 25 syndromes and as a non-syndromic entity (see below) and hypertelorism is recognised in more than 100 syndromes.12,13 It must be emphasised that although both GCPS and PHS are clinically distinct, both have a wide range of severity and it is incorrect to assert that one disorder is more severe than the other. This misconception was fostered because early reports of PHS were unusually severe,14 whereas early reports of GCPS were relatively mild.15 The recognition that the two phenotypes each manifested a full range of severity meant that they could not be placed on a single continuum of severity and that other mechanisms must be invoked to explain the phenotypes. This led to the pursuit of the bifunctional model, as delineated above.
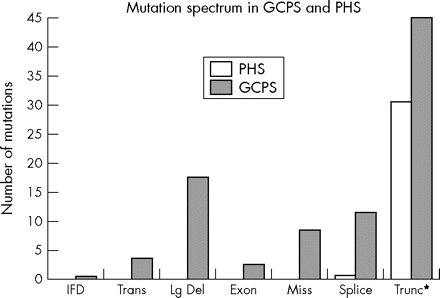
In spite of this ready clinical distinction of the two disorders, an argument was made that the allelism dictated that PHS and GCPS should be considered as a phenotypic continuum and should together be redesignated as GLI3 morphopathies.16 However, it is now clear that GLI3 mutations correlate with the two phenotypes on two levels. First, the classes of mutations that cause PHS and GCPS are distinct. The mutations that cause GCPS include translocations, large deletions, frameshift, missense, nonsense, and splice site mutations (fig 1). In contrast, the mutations that cause PHS are nearly completely limited to frameshift and nonsense mutations.17 Second, the position of the frameshift mutations among patients with the two disorders is distinct. Frameshift and nonsense mutations that are 5′ to, or include, the zinc finger encoding domain of the protein uniformly cause GCPS (35/35 known nonsense and frameshift mutations in this region, fig 2). Frameshift and nonsense mutations between the 3′ of the zinc finger domain and codon 1161 generally cause PHS (31 of 39 known nonsense and frameshift mutations). This correlation of the position of these two classes of frameshift mutations was predicted soon after the causation of GLI3 was established, based on an evolutionary comparison of the Drosophila ci homologue, as described above.10,18 Intriguingly, a third group of frameshift and nonsense mutations (12 of 12 frameshift and nonsense mutations) has been identified 3′ of codon 1161 and these mutations are uniformly associated with GCPS. In addition, there is a suggestion that this third group of patients may be less likely to manifest hypertelorism, but this has not been confirmed.17
Correlation of mutations and phenotype for the GLI3 gene. The classes of mutations that cause Greig cephalopolysyndactyly syndrome (GCPS) and Pallister-Hall syndrome (PHS) are distinct. Note that PHS is only caused by frameshift and nonsense mutations (plus one splice mutation, see text) whereas GCPS is caused by all types of mutations.
Correlation of mutations and phenotype for the GLI3 gene. Within the class of frameshift and nonsense mutations, there is a correlation of mutation position and phenotype. Note that all mutations in the 5′ part of the gene cause Greig cephalopolysyndactyly syndrome (GCPS) as do all of the mutations in the 3′ end of the gene. In the central part of the gene, nearly all of the mutations cause Pallister-Hall syndrome (PHS) and a few cause GCPS (adapted from Johnston et al17).
Thus the pleiotropy of GLI3 has several mechanisms. The first is the common mechanism whereby a gene product is expressed in multiple tissues during development as part of a genetic regulatory pathway that performs similar functions in various tissues or organ systems. The second is unusual and involves the bifunctional nature of a transcription factor whose dual activation and repression functions regulate apparently distinct developmental processes and generate distinct arrays of malformations by this mechanism.
VARIABLE EXPRESSIVITY I: THE RANGE OF SEVERITY IN GCPS
Clinical research into GCPS and PHS has provided additional insights regarding genotype–phenotype correlation. It was recognised decades ago that patients with GCPS had a mildly increased risk of developmental delay and mental retardation.3,15,19 It is always challenging to counsel families on prognosis when a disorder is associated with a mildly increased risk of a disabling or severe complication. It was also recognised that patients with severe GCPS overlapped clinically with those affected by the acrocallosal syndrome (ACLS; OMIM 200990, table 1).20,21 This overlap has substantial implications and makes it difficult to provide accurate prognoses and recurrence risks (ACLS is associated with severe mental retardation and seizures and is inherited in an autosomal recessive pattern). To investigate this conundrum two groups recognised the phenotypic overlap of these disorders and tested the hypothesis that some patients diagnosed with ACLS were instead affected by GCPS.22,23 The specific hypothesis was that some of these patients had a contiguous gene syndrome that included GLI3 within the deletion (thus causing the patients to have polysyndactyly, hypertelorism, and macrocephaly) but the patients were also deleted for various genes surrounding GLI3 and it was the deletion of these contiguous genes that caused the mental retardation and seizures.23,24 This syndrome has been designated as the GCPS contiguous gene syndrome (GCPS-CGS) to distinguish it from more typical GCPS and ACLS.23
One intriguing case report showed that a phenotype resembling acrocallosal syndrome resulted from a c.2800G→C GLI3 substitution, which predicts p.A934P. This suggested that it is possible for GLI3 mutants to generate dominant negative alleles that have distinct modes of action from those of the truncated GLI3 repressor forms which commonly result from truncations 3′ of the zinc finger domain that otherwise cause PHS. The analysis of additional patients with this phenotype should shed light on whether this particular patient is an example of allelic heterogeneity or of modifiers.
From these data it is clear that the spectrum of GCPS overlaps substantially with ACLS and that patients with these findings should undergo testing to preclude a contiguous gene deletion that includes GLI3 followed by mutation scanning of GLI3 before recurrence risks for autosomal recessive ACLS are provided to the family.
VARIABLE EXPRESSIVITY II: THERE IS NO SUCH THING AS A SINGLE GENE DISORDER
Although these insights into the GCPS contiguous gene syndrome explain much of the variability of the GCPS spectrum, it is not complete. In general, clinicians can counsel patients that in familial cases of PHS or GCPS, the severity of the disorder in future affected children is likely to be similar to that of the existing affected family members (excluding mosaic founders). There is a modest probability that the phenotype will be markedly more or less severe than that of the existing family members. The related issue of penetrance is germane here. There has been a single case of a family affected by GCPS with a documented occurrence of non-penetrance.22 While the complete details of this apparently non-penetrant patient were not reported, it is probably prudent to incorporate a high, but not 100%, penetrance for GCPS. Some of the variable expressivity of GCPS may be explained by allelic heterogeneity. However, there is no apparent correlation of mutation position and the severity within either the GCPS or the PHS groups, other than the suggestion that the 3′ frameshifting GCPS mutations may cause hypertelorism less often than the 5′ frameshifting mutations.17 Overall, the data are consistent with a loss of function or haploinsufficiency mechanism of GCPS and a gain of function mechanism for PHS. Therefore, one must look elsewhere to explain this variability. One potential source of such variability lies within GLI3. It is possible that variants in GLI3 regulatory elements may contribute to this variability. For example, if an otherwise “moderate” severity GCPS mutation was in trans with an allele that had a lower than average overall expression level, this would be expected to produce a more severe GCPS phenotype as the function of the two alleles may be additive. One would predict that the occurrence of a PHS mutation in cis with such a low expression variant would mitigate the severity of the primary mutation. Similar effects could result from splicing efficiency variants, as has been shown for cystic fibrosis transmembrane conductance regulator (CFTR).25 If these expression or splicing efficiency variants were in cis with the mutation, one would predict that the severity of the phenotype would breed true. Conversely, to the extent that such variants existed on the trans allele, one would predict that the variation would vary within families. The data to date indicate that the intrafamilial variability is less than the interfamilial variability,26 suggesting that both may be operative. In addition, it is likely that variants in other genes also contribute to the variability. Thus both phenotypes caused by GLI3 mutations show substantially variable expressivity and this variability cannot be explained by allelic heterogeneity, demonstrating that that an apparently simple single gene disorder is in fact genetically complex.
VARIABLE EXPRESSIVITY III: WHAT IS THE BOUNDARY SEPARATING SYNDROMIC FROM NON-SYNDROMIC POLYDACTYLY?
The polydactyly of GCPS is remarkably variable within families and even within a single individual when contralateral limbs are compared. In some cases, there is little in the way of preaxial duplication, and some patients manifest only a broad great toe, and it is important to emphasise that there are no objective standards for this assessment. As mentioned above, the most common form of upper limb polydactyly in GCPS is postaxial, but some patients have preaxial duplication in the hands. Even among familial cases of GCPS, it is clear that some affected persons have eudactylous limbs and, as mentioned above, there is a likely occurrence of non-penetrance. The situation becomes even more difficult when it comes to the craniofacial anomalies. Again, when evaluating affected familial cases of GCPS, it is clear that the craniofacial manifestations are markedly variable. Some affected relatives of patients who have clear facial dysmorphic features of GCPS (hypertelorism, macrocephaly, and a prominent forehead) have subtle or no craniofacial manifestations. This can be objectively assessed in the case of hypertelorism and macrocephaly—although it must be borne in mind that macrocephaly is a common variant and can segregate independently in families (OMIM 153470).
The situation in PHS is different from that in GCPS. In GCPS, the non-limb anomalies are easy to recognise and are clearly absent in some patients. In PHS, the non-limb anomalies are difficult to recognise because they are internal and often asymptomatic. The hypothalamic hamartomas of PHS most often cause no symptoms, although some patients can present with life threatening hypopituitarism (which is either secondary to or associated with the hamartoma) or seizures. This lesion is difficult to recognise on ultrasound (prenatal or postnatal) or computed tomography, and can only reliably be recognised on magnetic resonance imaging (MRI).27 Bifid epiglottis was present 15 of 26 patients with PHS, but all 15 were asymptomatic.28 Thus many patients with PHS could erroneously be considered to have non-syndromic polydactyly if they are not thoroughly evaluated.
In this light it is challenging to evaluate reports on “non-syndromic” polydactyly associated with GLI3 mutations. Clinical details included in these case reports are typically sparse and in some cases it is clear that the necessary evaluations were not carried out. For example, in the report by Radhakrishna et al,16 families UR014 and UR015 presented with apparent postaxial polydactyly and were found to have a mutation that is similar to that seen in patients with PHS. The few radiographs shown confirm postaxial polydactyly but we do not know if other members of the family may have had central polydactyly. While it is possible that this family has non-syndromic polydactyly (type undetermined), cranial MRI was not done. No patients from this family were evaluated for bifid epiglottis. Again, as most people with these anomalies would be expected to be asymptomatic if they had these findings, it is possible that these individuals have syndromic polydactyly or perhaps mild PHS. Similarly, a recent case report by Fujioka et al claims to describe a patient with non-syndromic preaxial polysyndactyly.29 Although the authors claim that the patient did not have hypertelorism, the photograph that was provided of the proband suggests that the craniofacial features were not normal, and details of the affected father were not presented. These two reports are examples of conclusions that are based on inadequate phenotyping that emanates from an incomplete understanding of the subtleties of these disorders. It should also be noted that the previously proposed model of pathogenesis (GCPS, PHS, and non-syndromic polydactyly) based on three classes of truncation mutations (successive from 5′ to 3′) was incorrect.10 Instead of there being three classes of phenotypes, there appear to be only two (GCPS and PHS), and this error was caused by this investigator’s failure to recognise the possibility that non-syndromic polydactyly was simply a mild variant of PHS instead of a distinct disorder.
While it is the case that endless arguments can be made about particular cases, the problem of non-syndromic polydactyly is formally insoluble. A negative scientific assertion cannot be proven. In this case it is impossible to prove that any given patient has no anomaly other than polydactyly. For PHS, hypothalamic hamartomas can be small and we have evaluated patients who had normal MRI on one occasion, whereas on a second occasion there was clearly a small hamartoma. In this case, the hamartoma was small enough for the shift of the MRI “slice” to be only a few millimetres, sufficient to allow or preclude identification of the mass. More generally, this debate is artificial. The syndromic/non-syndromic argument is an attempt to apply a categorical discrimination to a continuous biological variable, as there is great clinical utility in these kinds of labels. They are useful to sort patients into categories that allow appropriate clinical monitoring and care. It is useful, valid, and important to distinguish GCPS from PHS, but the distinction of syndromic versus non-syndromic polydactyly associated with GLI3 mutations is less useful biologically, even though the clinical label may still be appropriate. However, to suggest that “non-syndromic” post axial (or central) polydactyly associated with a GLI3 truncation mutation in the middle of the gene is biologically distinct from mild PHS is pointless. As noted above, the PHS and GCPS phenotypes are easy to distinguish, breed true, have markedly different prognoses and management, and have a distinct mutational spectrum and distinct pathogenic mechanisms.
THE MISSING MUTATIONS: YOU ONLY SEE WHAT YOU KNOW AND YOU CAN’T KNOW WHAT YOU DON’T SEE
The great majority of mutations in GLI3 are loss of function mutations in patients with GCPS and truncation mutations in patients with PHS. There are strikingly few reported missense mutations in the gene. This is almost certainly an artefact of the process of discovery that we and other groups have followed. By ascertaining patients on the basis of a few specific phenotypes, we have discovered useful and important molecular correlates of those phenotypes. However, we know next to nothing about the full range of phenotypes that may be caused by mutations in GLI3. This is because we do not know what other phenotypes could be caused by point mutations in the gene and we therefore cannot ascertain such patients. This problem is difficult to solve. To the extent that multiplex families with other phenotypes can be identified and linked to 7p13, GLI3 can be tested as a candidate. However, such families will be rare and sporadic cases are difficult to approach by this method. Instead, we will need to develop high throughput techniques to allow large numbers of genes (including GLI3) to be assayed in large cohorts of patients with a variety of phenotypes. This model-free approach is necessary to break through the limitations of current methods.
CONCLUSIONS
The phenotypes caused by mutations in GLI3 are diverse, discrete, variable, and pleiotropic. The mutations in GLI3 that cause PHS and GCPS correlate with the phenotypes on two levels: many types of inactivating mutations cause GCPS, whereas PHS is caused almost exclusively by truncation mutations in the middle third of the gene. This mutational correlation is supported by in vitro and animal model experimentation showing that the truncation mutations correlate with the post-translational regulation of the gene, which is accomplished by proteolytic processing to give GLI3 both a transcriptional repressor and activator effect. Thus GLI3 is a bifunctional transcriptional switch and these attributes correlate with the phenotype. The PHS and GCPS phenotypes caused by GLI3 mutations are qualitatively distinct, but both encompass a wide range of severity that may include non-syndromic polydactyly, although the data are incomplete in this regard. Further research is necessary to understand the full range of phenotypes caused by mutations in this complex and fascinating gene.
Acknowledgments
The research work performed by LGB and described here was supported by Intramural Research funds of the National Human Genome Research Institute, National Institutes of Health.
REFERENCES
Footnotes
-
Conflicts of interest: none declared